Genetics of obesity
 From Citizendium - Reading time: 11 min
From Citizendium - Reading time: 11 min
Obesity, a condition of excessive fat accummulation, is a major risk factor for premature mortality. While environmental influences (including the ease of access to high energy palatable foods and sedentary lifestyle) cannot be neglected, there is much focus on the genetics of obesity to explain variations in individual susceptibility to adiposity. Although several genes have been linked to obesity, they are limited to monogenic causes. Many of these are mutations of proteins in the “leptin pathway” which has an important role in energy balance. Each of those represented in the diagram has been shown to cause obesity, most often through twin studies.
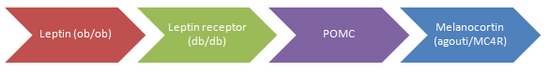
In all cases (except for MC4R), mutations are rare in the human population. However these findings suggest there can be more common multifactorial influences on susceptibility to adiposity which are likely to be involved in similar pathways.
Population genetics[edit]
Pima Indians[edit]
The Pima Indian population once resided in the deserts of Mexico where resources were scarce, but some migrated and communities now exist in Arizona, USA. Those who settled there were presented with a relative abundance of nourishment and exhibit extremely high levels of obesity and type 2 diabetes, whereas other communities of Pima do not. It has been proposed that genetic factors have protected this population in the past and allowed them to withstand conditions of deprivation, though if exposed to greater levels of food intake they are more susceptible to developing obesity as a result. This is known as the 'thrifty gene hypothesis'. [8]
Thrifty gene hypothesis[edit]
The thrifty gene hypothesis, first proposed by Neel in 1962, states that a genotype that stores energy more efficiently in times of food abundance would have been advantageous to our ancestors to survive times of food shortage.[1] It is suggested that this genotype has been naturally selected through years of food shortage but in modern society has become a source of health problems. These days, food is almost always easily available, so those with the thrifty phenotype are in constant 'food storage mode', preparing their bodies for a period of food shortage that never comes. This, it is suggested, is the cause of the widespread prevalence of obesity and diabetes mellitus type 2 in the developed world.
Recently this hypothesis has been challenged. JR Speakman, in a review for the International Society of Diabetes Vascular Disease, suggested that not enough significant famines have occurred in human populations, and that mortality during these famines would not have been sufficient to lead to the levels of natural genotype selection that the thrifty gene hypothesis implies. Mortality patterns also do not fit with the hypothesis, because deaths were often not due to starvation but to disease, so the thrifty phenotype would not have been particularly advantageous and those age groups incurring the highest mortality rates would have been the very young and very old, and not those of reproductive age so gene selection for future generations would be unaffected. Whether the thrifty geneotype explains some of modern day obesity remains unclear as records of famine and mortality rates in history are not always well kept.
Polynesian Populations[edit]
Polynesia is a subregion of Oceania, encompassing more than 1000 islands over the central and southern Pacific Ocean. Polynesians share language, culture, beliefs and other features of society. The populations are interesting for study because of the relatively conserved gene pool, the concept of modernisation, and the migration of Polynesians to other countries. Polynesian populations have high rates of type 2 diabetes and obesity.[2] However, increased rates of other obesity-associated problems such as metabolic syndrome and dyslipidaemia are not observed.[3] Finally, a study in several Oceanic populations did not support Neel's thrifty gene hypothesis: population frequencies of common FTO polymorphisms displayed no significant association with BMI.[4]
Monozygotic Twin Studies[edit]
Twin studies can be very useful for investigation of the gene-environment interactions of obesity [5] Using twins shows whether the already known inter-individual effects of changes in energy balance or dietary interventions are due to genetic factors. For example, exposing monozygotic twins to positive energy balance/overfeeding lets us investigate whether differing sensitivities in individuals gaining fat when exposed to positive energy balance is dependent on genotype. Variables in phenotype measured in the study included; body weight, body composition, fat distribution, abdominal visceral fat and resting metabolic rate. The findings after overfeeding/exercise in the twins conveyed clear intrapair resemblance and variation between different twin pairs. This suggested that differences in susceptibility of overeating must be controlled mainly by genetic factors, though the exact genes involved are not known.[6]
Using monozygotic twin studies also allows measurement of the relationship between dietary factors and body fat independent of genetic factors. [7] Both diet and genetic influences are thought to influence body fat. Using monozygotic twin studies it is possible to investigate the effect of diet on body fat independent of genotype. This lets us analyse the extent to which genetics is involved in influencing body fat, as no relationship between dietary fat and body fat was found in middle-aged women in a study conducted by Samaras et al [8], indicating that diet in determining total body fat may have been overestimated in the past and genetic factors may be more influential.
Monogenetic forms of Obesity[edit]
Mendelian Disorders[edit]
Obesity is a major clinical feature (but not the dominant feature) of some Mendelian disorders for which genetic mutations have been found. [9] Prader-Willi syndrome is autosomal dominant and characterised by obesity, reduced foetal activity, mental retardation and hyperphagia which develops between the age of 1 and 2. Around three quarters of patients have a deletion on chromosome 15 (15q11-q13) and the remaining have maternal disomy. Albright hereditary osteodystrophy is a rare autosomal dominant disorder where patients are obese, short, mentally retarded and have subcutaneous calcifications. AHO is due to parental imprinting of mutations in the GNAS1 gene. [9] The mutations for these have been established but, despite attempts, there has been no link between mutant genes and disrupted energy balance. It is likely that the underlying genetics that cause obesity in these Mendelian disorders are different to those in common forms of obesity, though further studies investigating genetic markers would need to be carried out. [9]
Single-gene disorders[edit]
Leptin
Leptin is a hormone secreted from adipocytes which allows information regarding energy stores to be transmitted to the brain, primarily the hypothalamus. It inhibits appetite by inhibiting orexigenic NYP/AgRP neurons in the arcuate nucleus and it stimulates metabolic rate. The levels circulating in the blood are proportional to the amount of adipose tissue.
A very small number of patients worldwide have been found to have leptin mutations. Montague et al. [10]investigated two morbidly obese hyperphagic child cousins with undetectable leptin. They were both found to be homozygous for deletion of a single guanine on codon 133, leading to a premature stop codon. The phenotype of these children is very like that of the ob/ob mice which are obese, hyperphagic, infertile and have hyperinsulinaemia. It was not possible to look into the reproductive effects of these children as they were pre-purbertal, but it was speculated leptin is also required to initiate puberty.
Farooqi et al. [11] treated these children with human recombinant leptin daily for a year, weighing them daily, carrying out DEXA scans, recorded food intake and measured other hormone levels such as insulin, thyrotrophin and gonadotrophins. They lost on average 1-2kg per month, their appetite decreased and all other hormones were normal. Their GnRH became pulsatile after 12 months indicating leptin may affect puberty onset.
Another mouse with a similar phenotype to ob/ob mouse is the db/db mouse. It has very high leptin and was found to have a leptin receptor mutation due to a premature stop codon which codes for a receptor without the intraceullar C terminal, which is necessary for tyrosine kinase activation. Only one family have been found to have a leptin receptor mutation which were homozygotes were hyperphagic, obese, growth retarded and had hypothalamic hypothyroidism. This indicates that the lack of receptors causes certain hypothalamic releasing factors to be impaired. [9]
POMC
POMC (pre-pro-opiomelanocortin) is cleaved by prohormone convertases to form the melanocortin peptides adrenocorticotrophin (ACTH) and melanocyte-stimulating hormones (MSH-alpha, beta and gamma), and also beta-endorphin, an opiod receptor ligand. Discoveries about the link between alpha-MSH and regulation of food intake led to interest in the association of POMC with obesity. Scientists predicted, through knowledge of the normal physiology of POMC and melanocortins, that the phenotype for POMC mutation would show obesity, pigmentation changes and ACTH deficiency. Two patients showing this phenotype were initially found and sequencing of their POMC genes revealed mutations that abolished POMC function, and therefore no melanocortin peptides could be detected in these patients. [12]
Many single nucleotide polymorphisms have now been identified that cause POMC deficiency syndrome, characterised as early-onset obesity, hyperphagia, hypocortisolaemia due to ACTH deficiency and often red hair with pale skin due to lack of ligand for melanocortin-1 receptors. However, pigmentation changes may be obscured by differing genetic backgrounds. Patients homozygous for POMC-null mutations exclusively exhibit the obesity phenotype but this is extremely rare. Heterozygous patients have been shown by analysis of family pedigrees to be more prone to obesity.[13]This has led to the suggestion that common obesity may be due to heterozygous genotypes for POMC mutations. In animal experiments, POMC-null heterozygous mice become obese on a high fat diet but remain slim on normal chow.[14] This indicates that environmental influences could affect whether heterozygous obesity genes result in the obesity phenotype. Extensive screenings of obese cohorts which don’t display other POMC-deficiency syndromic effects have not been conclusive about whether POMC mutations cause common obesity. In one study, only 6 out of 601 obese patients showed mutations in the non-coding regions of the POMC gene which could be linked to their obesity. [15]This suggests POMC mutations may not significantly contribute to common obesity.
MCR4 Melanocortins act on melanocortin receptors; the main subtype of receptor that is involved in feeding energy balance is melanocortin-4 receptor (MC-4R). Mice homozygous for the MC-4R-null mutation show severe obesity and homozygotes show increased weight compared to wild-type mice but less severe. [16]Many human cases of heterozygous MC-4R mutations have now been identified and sequenced. In a large cohort of 2000 severly obese subjects 5-6% were found to have mutations of the MC-4R. More recent studies showed that 1-2.5% of subjects with a BMI over 30 had a MC-4R defect. These results suggest that MC-4R mutations are the most common genetic cause of obesity to date.[17]
carboxypeptidase E
Genetic disruption to POMC and other prohormone post-translational processing has been linked to obesity due to lack of production of MC-4 ligands. Carboxypeptidase E (CPE) is an enzyme which processes prohormones. A naturally occurring mutation in the gene encoding CPE causes obesity as in the fat/fat experimental mouse model. Being homozygous for the fat mutation results in obesity, exogenous insulin-sensitive hyperglycaemia, hyperproinsulinaemia and infertility. CPE is a receptor on the golgi membrane for prohormones like POMC to bind, which signals budding off of a prohormone-containing vesicle.[18]Cleavage of prohormones in these vesicles is then mediated by prohormone convertases (Pc-1 and Pc-2) which are differentially regulated according to cells type to respond to nutritional and hormonal signals.[19]Mice transgenically expressing an inactive form of PC-1 show an obese phenotype.[20] This presents pharmacological potential to target specific steps in prohormone packaging and cleavage to aid weight loss.
FTO Through modern genotyping technology, genome-wide association studies are now possible, allowing researchers to correlate genetic variants with particular traits on massive scales. It is through this method that polymorphisms in the FTO gene were identified as having significant influences on BMI in Caucasian individuals, in a study of a population of Sardinians by Scuteri et al (2007) [21]. The researchers examined genetic data from over 4,000 subjects and found strong associations between several Short-Nucleotide Repeats (SNPs) in the first intron of the FTO gene and BMI, hip circumference and total body weight. While shown to have only a modest influence of BMI; with each risk allele increasing body weight by around 1.3 to 2.1 kilograms, the frequency of risk allele expression is high in the European population at around 63% [22]. This is therefore a discovery with clinical relevance and represents a common genetic determinant of obesity.
The normal biological purpose of the FTO gene is not well understood, but it is believed to play a role in central appetite regulation. FTO mRNA is expressed in the feeding centres of the hypothalamus, including the supraoptic, ventromedial and arcuate nuclei. [23] Individuals carrying the risk alleles for FTO will not inevitably become obese. Nevertheless, the discovery of the FTO gene as a predisposition for increased body weight provoked excitement and speculation that further, commonly expressed polymorphisms may exist which account for differing susceptibilities to obesity amongst different individuals.
Polygenetic forms of Obesity[edit]
Cases of obesity caused by single-gene polymorphisms are extremely rare, but subtle interactions between the polygenic variants in a range of genes are much more common. A polygene can only be identified and validated by showing that such an allele occurs more frequently in obese patients than those that are not obese. Even once identified, each single polygene makes a tiny contribution to an obese phenotype [21]. The nuances within obese phenotypes, with the additional effects of the environment, make determining the genetic basis of obesity extremely difficult.
Candidate genes are selected on the basis of their involvement in pathways of energy balance or adipose tissue biology. Positive associations with an obese phenotype have been observed for more than 70 genes, but studies are not consistent and many associations have not been replicated in more than one study or population. Both transgenic knockout mice studies and human studies identified the genes for leptin and its receptor as candidate genes: polymorphisms in these coding regions showed positive associations with body weight, weight loss and BMI. Disappointingly, despite leptin’s important role in appetite regulation, only a handful of mutations/single-nucleotide polymorphisms have been functionally associated with obesity. Association studies between obese phenotypes and other key proteins (POMC, AgRP, MC4R, NPY) are limited and require replication.
A different approach is to use genome-wide scans to identify quantitative trait loci (QTL)s , and eventually focus on specific genes within the chromosomal region. If strong evidence is found in an identified QTL, more sensitive screening of that region is carried out with a denser map of markers. Strong evidence for a QTL influencing obesity was found on chromosome 2p22 and has been linked with leptin and adiponectin levels in further studies [22]. Other QTLs with associations found include chromosomes 3q27, 5p-cen, 6p21-22, 7q, 10p, 11q23-24, 17p12, 2and 0q11-13. New linkage methods have looked at pairwise correlation analyses and showed strong correlations between 10q and 20q. Other studies have looked into imprinted genes and their effect on obesity.
A study using an outbred animal model of obesity (M16 mice) suggested that such a model could be used to look at gene discovery and the pathways involved. The phenotype in mice mirrors the development of obesity in humans [23]. A 1997 study considered the homology between human and animal models, with regards to estimating the number, location and effect of QTLs controlling obesity. The study looked forward to consider comparable “obesity maps” based on animals that were applicable in humans [24]. The difficulties in reliably demonstrating genes involved in obesity are again shown in a paper describing common polygenetic obesity. Unconfirmed associations may be due to inadequate sample sizes or inadequate examination of variation within candidate genes [25].
References[edit]
- ↑ Neel JV (1962) Diabetes mellitus: a 'thrifty' genotype rendered detrimental by 'progress'? Am J Hum Genetics 4:352-3
- ↑ McGarvey ST (1991) Obesity in Samoans and a perpective on its etiology in Polynesians. Am J Clin Nutr 53:1586S-94S PMID 2031491
- ↑ Cournil A et al. (2007) CALDIA Study Group. Paradoxical relationships between anthropometric variable and phenotypic expression of the metabolic syndrome in nondiabetic Polynesians of New Caledonia Diabetes Care 30:1909-11 PMID 17446532
- ↑ Ohashi J et al. (2007) FTO polymorphisms in oceanic populations J Hum Genet 52:1031-5 PMID 17928949
- ↑ Bouchard C et al. (1990) Using MZ twins in experimental research to test for the presence of a genotype-environment interaction effect. Acta Genet Med Gemellol 39:85-9 PMID 2392894
- ↑ Bouchard C, Tremblay A ( ) Genetic influences on the response of body fat and fat distribution to positive and negative energy balances in human identical twins J Nutr 127:943S-7S PMID 9164270
- ↑ Hasselbalch AL et al. (2010) Associations between dietary intake and body fat independent of genetic and familial environmental background. Int J Obes 34:892-8 PMID 20125102
- ↑ Samaras K et al. (1998) Genes versus environment. The relationship between dietary fat and total and central abdominal fat Diabetes Care 21:2069-76 PMID 9839096
- ↑ 9.0 9.1 9.2 9.3 Loos RJ, Bouchard C (2003) Obesity-is it a genetic disorder? J Intern Med 254:401-25 PMID 14535962
- ↑ Montague CT et al. (1997) Leptin deficiency is associated with severe early-onset obesity in humans Nature 387:902-8 PMID 9202122
- ↑ Farooqi S et al. (1999) Effects of recombinant leptin therapy in a child with congenital leptin defiency New Eng J Med 341:879 PMID 10486419
- ↑ H Krude et al. (1998) Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans Nature Genetics
- ↑ Farooqi S et al. (2006) Heterozygosity for a POMC-null mutation and increased obesity risk in humans Diabetes 55:
- ↑ Challis BG et al. (2004) Mice lacking pro-opiomelanocortin are sensitive to high-fat feeding but respond normally to the acute anorectic effects of peptide- YY3-36 PNAS
- ↑ H Krude et al. (2003) mutations in the proopiomelanocortin gene Ann NY Acad Sci
- ↑ Huszar D et al. (1997) Targeted disruption of the melanocortin-4 receptor results in obesity in mice Cell 88:131–41
- ↑ Farooqi S, O’Rahilly S ( ) Genetics of obesity in humans Endocrine Rev 27:710-8
- ↑ Cool DR et al. (1997)Carboxypeptidase E is a regulated secretory Pathway sorting receptor: genetic obliteration leads to endocrine disorders in Cpefat mice Cell 88:73–83
- ↑ Leona Plum et al. The obesity susceptibility gene Cpe links FoxO1 signaling in hypothalamic pro-opiomelanocortin neurons with regulation of food intake Nature Med 15:1195-201 (2009)
- ↑ Lloyd DJ et al. (2006) Obesity, hyperphagia and increased metabolic efficiency in Pc1 mutant mice Hum Mol Genet 15:1884–93
- ↑ Hinney & Hebebrand (2008) Polygenic Obesity in Humans. Obesity Facts 1:35-42 PMID 20054160
- ↑ O'Rahilly et al. (2003)Human obesity - lessons from monogenic disorders Endocrinology 144: 3757-64 PMID 12933645
- ↑ Allan et al. (2004)The M16 mouse: an outbred animal model of early onset polygenic obesity and diabesity Obes Res 12:1397-407 PMID 15483204
- ↑ Pomp D (1997) Genetic dissection of obesity in polygenic animal models Behav Genetics 27:285-306 PMID 9519558
- ↑ Walley et al.(2006) Genetics of obesity and the prediction of risk for health Human Mol Gen 15:R124-30 PMID 16987875
 KSF
KSF