Jay Dunlap
Topic: Biography
 From HandWiki - Reading time: 18 min
From HandWiki - Reading time: 18 min
Jay Dunlap | |
|---|---|
| Born | May 9, 1952 Ludlow, Massachusetts |
| Alma mater | University of Washington, BS & BS 1974 Harvard University AM 1975 Harvard University PhD 1979 University of California, Santa Cruz PPhD 1979-1983 |
| Known for | Neurospora Circadian Rhythms |
| Awards | Honma International Prize For Biological Rhythms Research(1991), Genetics Society of America Robert L. Metzenberg Award(2005) George W. Beadle Medal, Genetics Society of America(2009), AAAS Fellow(2010), Fellow of the American Academy of Microbiology(2010), National Academy of Sciences(2009) |
| Scientific career | |
| Fields | Biochemistry, Genetics, Chronobiology |
| Institutions | Department of Genetics, Dartmouth Medical School |
| Website | geiselmed |
Jay Dunlap is an American chronobiologist and photobiologist who has made significant contributions to the field of chronobiology by investigating the underlying mechanisms of circadian systems in Neurospora, a fungus commonly used as a model organism in biology, and in mice and mammalian cell culture models. Major contributions by Jay Dunlap include his work investigating the role of frq and wc clock genes in circadian rhythmicity, and his leadership in coordinating the whole genome knockout collection for Neurospora. He is currently the Nathan Smith Professor of Molecular and Systems Biology at the Geisel School of Medicine at Dartmouth. He and his colleague Jennifer Loros have mentored numerous students and postdoctoral fellows, many of whom presently hold positions at various academic institutions.
Early life and education
Born in Ludlow, Massachusetts on May 9, 1952, Jay Dunlap grew up in York, Pennsylvania as the third of four children.[1] Dunlap became interested in biochemical oceanography during a high school summer program and decided to pursue this interest in college. He graduated with a B.S. in oceanography and a B.S. in chemistry from the University of Washington in 1974.[1]
Dunlap originally planned to pursue oceanography in his graduate studies. However, after meeting with John Woodland Hastings, who studied the circadian regulation of bioluminescence in marine organisms, Dunlap decided to study biology in graduate school at Harvard University. While studying with Hastings, Dunlap changed his field of study to circadian biology.[1][2]
Career and research
For his postdoctoral fellowship, Dunlap attended the University of California, Santa Cruz and started working with Jerry Feldman, who had successfully isolated clock gene mutants in Neurospora that have abnormally long or short circadian-oscillation periods. Dunlap was unable to clone frequency, a gene that has an important role in the transcription-translation negative feedback-loop (TTFL) that drives circadian rhythms in Neurospora, as the Santa Cruz lab did not have the molecular tools necessary to study Neurospora's molecular biology in depth.[3] Dunlap learned basic molecular techniques as he worked alongside fellow biology graduate students in other labs. At one point, Dunlap worked with Harry F. Noller, a renowned biochemist whose lab had "unofficially adopted" Dunlap.[3]
In 1984, Dunlap secured a junior faculty position at the Department of Biochemistry at Geisel School of Medicine at Dartmouth. He became a professor of Biochemistry in 1994 before being named the Inaugural Chair of the Department of Genetics in 1999. In 2010, Dunlap was named Nathan Smith Professor, and in 2016, he was appointed inaugural chair of the Department of Molecular and Systems Biology which subsumed Genetics and other departments.[4]
Working closely with Jennifer Loros' laboratory, Dunlap's research has primarily focused on the molecular basis of circadian rhythms using Neurospora as a model system to further understand the mammalian circadian clock. Although clock gene mutations were also identified in Drosophila and Chlamydomonas,[1] Dunlap studied Neurospora in his postdoctoral work, as a wider array of biochemical and genetic tools were applicable to the species at the time.[3] Neurospora was a simple model organism and a powerful tool to study molecular genetics; its then-unknown molecular clock presented a great opportunity for exploration.[4]
Identifying Neurospora clock components and mechanisms
Based on the work of Dunlap and others, clock genes are now understood to encode proteins that participate in a self maintaining negative feedback loop: transcriptional activators drive expression of specific clock gene mRNAs, which are translated into clock proteins, which enter the nucleus and act to depress the activity of the transcriptional activators driving the expression of the clock genes.[5] However, clock genes were not yet cloned when Dunlap began his research as an assistant professor in 1984. Dunlap correctly predicted that single cells, including mammalian cells, can act as autonomous oscillators with their own intrinsic circadian rhythms.[6]
Dunlap deciphered the circadian system by framing and addressing three problems in cellular metabolism:
- How is the clock put together: what are the gears and cogs, how do they mesh, what regulates them, and how do they regulate one another so the collective output is a molecular/biochemical cycle with all the circadian characteristics?
- How do abrupt and transient changes in the environment, chiefly ambient light or temperature, reset the phase of the clock and align the internal clock of an organism with the external time?
- How is an intracellular molecular cycle used to regulate the behavior of the cell?[3]
Prior to the adoption of transcriptional reporters such as luciferase, studies of the Neurospora circadian clock utilized the rhythmic development of asexual spores (conidia), assayed using a race tube.[7] Conidial production peaks in the subjective night—a behavioral phenotype lacking in arrhythmic strains. During her graduate work, Jennifer Loros observed mutant frq9 as a recessive, arrhythmic, and phenotypically null allele at the frq gene.[8] Her observation, combined with the ability to transform Neurospora with exogenous DNA, provided the basis for a novel strategy to clone frq, namely by transformation-based rescue of the null mutant behavioral phenotype. Utilizing a bidirectional chromosome walk beginning at oli, a gene on the same linkage group as frq, Dunlap and colleagues walked over 200kb across frq.[3] The location of frq was verified in 1986 through transformation of cosmids into frq9 and by rescuing the circadian rhythm. frq was thus the second clock gene to be cloned, following Drosophila per. Furthermore, the lab manually sequenced roughly 9kb and conducted transcript mapping on the frq genomic region; the results were published in Nature in 1989.[3] In subsequent work, Dunlap and colleagues showed that frq was rhythmically expressed and were able to manipulate the expression of frq sufficiently to create a null mutant. They implemented a system in which a heterologous promoter—induced in a manner that did not affect the clock—could be used to drive regulated expression of frq. Using this system, they demonstrated that the product of frq acted to repress its own synthesis; it was autoregulatory. Dunlap and colleagues observed that the continual over-expression of frq resulted in arrhythmicity, and they defined the phase of clock's rhythm to be the time at which the cell returned to normal expression levels of frq. They concluded, in a Science article in 1994, that the core pacemaker of the Neurospora clock is regulated via negative feedback by clock proteins, and frq determines its own expression through auto-regulation via negative feedback, demonstrating that intracellular, auto-regulatory negative feedback is the basis of a circadian oscillator.[9][10]
Dunlap's work on the auto-regulatory mechanism included modeling the circadian clock's negative feedback loop and discovering the roles and connections between activators (which he identified as proteins with PAS domains) and repressors (products of the clock genes).[11] Additionally, Dunlap demonstrated the role of protein phosphorylation in the clock mechanism and has done research involving the role of these proteins (namely Casein Kinase 2) on the temperature compensation mechanism. In 2009, Dunlap and colleagues showed that the FRQ protein is phosphorylated at over 100 sites in a highly reproducible and time-of-day-specific manner[12] and that casein kinase 2 establishes and maintains temperature compensation within the circadian clock.[13] Four years later in 2013, Dunlap and colleagues found that FRQ is an Intrinsically Disordered Protein whose stability is determined by its interaction with partner protein FRH. Additionally, Dunlap and colleagues discovered that the daily phosphorylation of FRQ governs its ability to interact with the proteins in the negative element complex.[14] The kinetics of these circadian processes, Dunlap discovered, are heavily influenced by progressive phosphorylation of FRQ.[15]
Mechanism of entrainment

After identifying frq as a clock gene whose product's abundance tends to be variable and rhythmic, Dunlap, Loros, and colleagues showed how environmental regulation of its expression led to understanding the molecular basis of circadian entrainment by light: through the induction of expression of frq by light.[16]
In 1995, Loros and Dunlap worked to uncover the molecular basis underlying how light resets the clock, a mechanism later shown in collaborative work with Hitoshi Okamura to be conserved in mammals.[17] The daily cycle in frq transcript levels, combined with the ability of light to acutely induce frq expression, explained light resetting (the advances and delays seen on a phase response curve). If light was provided and induced frq-mRNA when it was rising to peak levels (late subjective night), light would rapidly bring frq-mRNA levels to peak values, resulting in an advance. If light induced frq-mRNA while its levels were falling (early subjective night), frq-mRNA would rapidly go back to peak levels causing a phase delay. The results of this investigation led to the conclusion that the light induction of frq is responsible for the phase-specific advances and delays observed in Neurospora and provided a general explanation for how the unidirectional response of a clock component to an environmental signal (light) could result in a bidirectional time-of-day specific clock response (advances or delays): the basis for circadian entrainment.[2] These experiments eventually led to the universal recognition of entrainment via light-induced changes in a specific variable of the circadian oscillator, later observed in Drosophila and mammals.
Identification of PAS-PAS heterodimers as activators in the circadian feedback loop
The mechanism through which frq is induced by light was unknown at the time that entrainment was explained, and studies aimed at identifying the proteins responsible for light-induction of frq led to the identification of White Collar-1 and White Collar-2 as components of the circadian activator complex.[18] Work by Giuseppe Macino had shown White Collar-1 to associate via PAS domains with White Collar-2 to create the White Collar Complex; Dunlap, Loros and colleagues showed how this heterodimeric complex is the transcription factor that acts in the dark to drive expression of frq, thereby acting as the activator in the circadian negative feedback loop. This observation associated specific biochemical activities, DNA binding and transcriptional activation, with known clock proteins, allowing the formulation of the oscillator as a single step transcription-translation negative feedback loop.[18] Later, in 1997, the first mammalian clock gene (CLOCK) was shown to encode a protein similarly having PAS domains and, later, to associate via PAS domains with a different protein, BMAL1, again forming a heterodimeric protein complex that acted as a transcriptional activator; similar proteins were identified in 1998 in Drosophila. This confirmed a common model for the transcription-translation negative feedback loops in fungi and animals: a positive element composed of two different proteins interacting via PAS domains drives expression of negative elements such as FRQ or PER that, in association with other proteins, represses the activity of heterodimeric activators: negative feedback.[19] These observations contributed to the naming of Circadian Rhythms as first runner up to Breakthrough of the Year in Science magazine in 1997.
Identification of a circadian photoreceptor
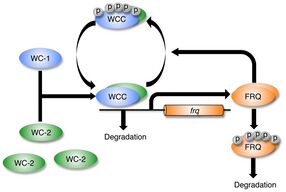
Although it was established that heterodimeric WC-1/WC-2 transcription factor was required for light-induction of frq, researchers believed that WC-1 and WC-2 did not have a direct role in the process of photoreception. WC-1/WC-2 transcription factor was instead assumed to be the final target of a signal transduction cascade initiated by the action of light on a distinct blue light photoreceptor. In 2002, Dunlap and colleagues biochemically studied WC-1/WC-2 in vitro to show that WC-1 bound FAD as a cofactor (also shown independently by Yi Liu), and analysis of binding to DNA by the WC-1/WC-2 complex showed that light resulted in a structural change in the heterodimer. The dose response and action spectrum for this in vitro structural change in WC-1 was FAD-dependent and matched the in vivo dose response and action spectrum for light-suppression of circadian banding determined by Briggs and colleagues in 1967. These findings revealed that WC-1 is a blue light photoreceptor and a circadian photoreceptor; the signal transduction cascade from photoreceptor to transcription factor happens all within the same protein.[21][15] WC-1 is the founding member for the family of blue-light photoreceptors common to all fungi.[22] Circadian photoreceptors were later identified in animals and green plants and shown to be distinct from WC-1.
Circadian output
In 1989, Dunlap's work with Jennifer Loros led to the first targeted screen for genes regulated by the circadian clock, paving the way for the systematic dissection of clock output pathways.[23] The term "clock-controlled genes" (CCGs) was coined in this study. CCGs are defined as genes whose level of expression is regulated by the circadian clock but whose activities do not impact the operation of the clock. Circadian control of gene expression is now widely thought to be the principal means through which clocks control the biology of the cells in which they operate. Subsequent work expanded the universe of CCGs in Neurospora, and later in mammalian cells,[24] and revealed the connection between the circadian and cell cycles in which the clock regulates the DNA damage response which, in turn, can regulate the clock.[25] The search for CCGs finally culminating in the complete description of the circadian transcriptome of Neurospora where as much as 40% of the genome is controlled on a daily basis by the clock.[26]
Studies on bioluminescence
Jay Dunlap’s graduate work at Harvard with J.W. Hastings focused on bioluminescence in the marine organism Gonyaulax. Their work uncovered the structure of Gonyaulax luciferin. After purifying luciferase, they determined that it was regulated through daily synthesis and destruction.[3] This was one of the first clock-regulated enzymes whose method of regulation was determined under experimental conditions. One part of the mechanism is that Gonyaulax produces luciferin and luciferase at night when the emitted light can be seen, while production of the substrate and protein decreases at dawn. The realization that a complete understanding of this biochemical process would also require a combined genetics approach led Dunlap to begin his study of the circadian clock of the Neurospora.[2]
Dunlap and colleagues later developed bioluminescence as a reporter for gene expression in Neurospora. Prior to the use of bioluminescence the only assay for rhythmicity in Neurospora was the daily cycle in asexual development (conidiation). As a result, strains bearing mutations that interfered with development could not be accurately assayed for rhythmicity. Dunlap, along with Jennifer Loros, Arun Mehra, and Van Gooch, adapted firefly luciferase for expression in Neurospora, thereby greatly expanding the ability to analyze strains.[7] frq-promoter-driven luciferase is an exquisitely sensitive reporter for the core oscillator and has been used show that developmental rhythms that do not require frq are not truly circadian,[27] and that daily phosphorylation of FRQ protein, but not daily turnover of FRQ protein, is required for closing of the negative feedback loop.[28] The novel method used by Dunlap and his colleagues to characterize and use the luciferase gene improved expression by 3 log orders and allowed for the correction of several errors in the Neurospora literature. Dunlap and Loros collaborated with Cassius Stevani to show that bioluminescence of the basidiomycete (mushroom) Neonothopanus gardneri is regulated by circadian rhythms through regulated expression of the luciferase, luciferin, and a required reductase.[29] N. gardneri is found growing beneath palms in the Amazonian forest and the nocturnal bioluminescence is believed to be used by the fungus to attract insects at night as an aid to spore dispersal.[30]
Technological advancements
Dunlap and his colleagues have contributed greatly to advancements in the use of technology within the field of molecular biology. These methodological advancements have had major implications for both fungal biology and chronobiology and their future directions. For example, Dunlap's lab developed the first gene replacement for Neurospora in 1991. These technologies as well as Dunlap's support greatly contributed to the sequencing of the Neurospora genome (which was accomplished in 2002). Subsequently, Dunlap and his team improved gene replacements. He spearheaded the push to knock out all 10,000 genes in the Neurospora genome and construction of a high-density single nucleotide polymorphism map. Finally, Dunlap revolutionized the role of luciferase expression by examining codon bias and is using its implications in Neurospora and other organisms.[7]
Present work
Dunlap continues to investigate the circadian clock, using Neurospora and other organisms, such as Aspergillus fumigatus.[31] As a result of the Neurospora crassa Genome Project,[32] the results of which were published in 2003, and the development of knockouts for every gene, which are stored at the Fungal Genetics Stock Center, Dunlap believes the molecular basis for the circadian clock of Neurospora may be the first to be completely understood. Due to the highly conserved nature of biological clocks, clock mechanisms have evolved relatively few times and are similar between species. Knowledge of Neurospora systems may lead to applications with relevance to human health. The circadian nature of cellular processes in humans may be leveraged to target cancerous cells more effectively and treat sleep abnormalities.
Dunlap is also interested in the interaction between biological clocks and metabolic processes. While circadian rhythms govern aspects of metabolism, metabolic products may feedback to an organism's internal clock.[33] This form of communication may prove to be an adaptive feature of biological clocks and enable beneficial responses to changes in environment. Additionally, Dunlap works with William Cannon and Jennifer Hurley to develop mathematical models describing circadian clock function. This effort will make use of statistical techniques to model both reactions occurring in metabolism and the overall clock.
Dunlap has also been involved in work examining the hierarchical network of transcription factors that govern circadian output. The core oscillator generates rhythmic activity of the heterodimeric circadian activator (WC-1/WC-2 or CLOCK/BMAL1), but the peak activity is restricted to one time of day. Thus, in Neurospora, the core oscillator that generates time creates rhythmic activity of the WC-1/WC-2 heterodimer that peaks in the morning. WC-1/WC-2 sits on top of a network of transcription factors where different tiers of regulators work together to act as a dynamic filter for time information, changing the morning peak activity of WC-1/WC-2 into a signal that can drive circadian gene expression at all times of day. A part of this is the transcription factor ADV-1.[34] This factor, found in Neurospora, responds to light and regulates genes involved in processes such as cell growth.
Recently, Dunlap looked into the evolutionary conservation of the circadian clock among species. Specifically, he found that proteins conserved in biological clock mechanisms among three species (Drosophila melanogaster, Neurospora crassa and Mus musculus) all exhibit high amounts of intrinsic protein disorder. Intrinsically disordered proteins do not have a stable secondary structure. Throughout the day, these proteins have different levels of disorder. The changing levels of disorder allow for a stable circadian rhythm. Dunlap concluded that because disordered proteins are so conserved among different species, the proteins must be essential for the control of the circadian rhythms across species.[35]
In his most recent work, Dunlap's lab examined regulators of the mRNAs encoding the Casein Kinase 1 protein; one such regulator is an RNA-binding protein translated from the prd-2 gene. They examined two mutations (created by inversion of a part of the prd-2 gene) and found that these mutations drastically affected Casein Kinase levels. These mutations caused circadian periods much greater than 24 hours. He and his colleagues genetically increased the Casein Kinase 1 levels and found that the period was restored when Casein Kinase 1 levels increased. They concluded that the circadian period is dependent on Casein Kinase 1 levels.[36]
Personal life
During Dunlap's time at Santa Cruz, one of the biology graduate students he met was Jennifer Loros. They forged a permanent relationship and were married on September 1, 1984. They have two children. When he is not conducting research, Dunlap enjoys gardening.[3]
Memberships, honors, and awards
Memberships
Jay Dunlap is currently involved with the following organizations:
- editorial board, Journal of Biological Rhythms (1994–2001; 2014–present)
- editorial board, G3: Genes, Genomes, Genetics (2011–present)
Previously, he has participated in:
- President, Society for Research on Biological Rhythms, (1998–2000)
- National Advisory Council for General Medical Sciences, (2000–2004, 2011)
- Founding Editor, Eukaryotic Cell (ASM Press), (2001–2011)
- co-Editor-in-Chief, Advances in Genetics (1995–2017)
Honors and awards
- 1980 Damon Runyon-Walter Winchell Fellowship
- 1983 National Research Service Award, NIH
- 1991 Honma International Prize For Biological Rhythms Research
- 1992 - 1997 Senior Scientist Award, National Institute of Mental Health
- 1998 MERIT (Method to Extend Research in Time) award, NIGMS
- 2005 (first) recipient of Robert L. Metzenberg Award, Genetics Society of America
- 2009 George W. Beadle Award, Genetics Society of America
- 2009 elected to the National Academy of Sciences, Genetics section
- 2010 elected fellow of American Association for the Advancement of Science
- 2010 elected to the American Academy of Microbiology
- 2013 fellow, Texas A&M University Institute for Advanced Studies
- 2017 PM Lecture, 29th Fungal Genetics Conference, Genetics Society of America
Key Publications
Research articles
- Loros, J.J.; Denome, S.A.; Dunlap, J.C. (1989). "Molecular cloning of genes under control of the circadian clock in Neurospora." Science. 243: 385–388. doi:10.1126/science.2563175 PMID 2563175
- Aronson, B.D.; Johnson, K.A.; Loros, J.J; Dunlap, J.C. (1994). "Negative feedback defining a circadian clock: autoregulation of the clock gene frequency." Science. 263(5153): 1578-84. doi: 10.1126/science.8128244 PMID: 8128244
- Crosthwaite, SK; Loros, JJ; Dunlap, JC. (1995). "Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript." Cell. 81(7): 1003-12. doi: 10.1016/s0092-8674(05)80005-4 PMID: 7600569
- Crosthwaite, S.K.; Dunlap, J.C.; Loros, J.J. (1997). "Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity." Science. 276(5313): 763-9. doi: 10.1126/science.276.5313.763 PMID: 9115195
- Liu, Y.; Merrow, M.; Loros, J.J.; Dunlap, J.C. (1998). “How temperature changes reset a circadian oscillator.” Science. 281: 825-829. doi:10.1126/science.281.5378.825 PMID 9694654
- Dunlap, J.C. (1999). “Molecular bases for circadian clocks.” Cell. 96: 271-290. doi:10.1016/S0092-8674(00)80566-8 PMID 9988221
- Froehlich, A.C.; Liu, Y.; Loros, J.J.; Dunlap, J.C. (2002). “White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter.” Science. 297: 815-819. doi:10.1126/science.1073681 PMID 12098706
- Baker, C.L.; Kettenbach, A.N.; Loros, J.J.;Gerber, S.A.; Dunlap, J.C. (2009). "Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora cirdadian clock." Cell. 34(3): 354-63. doi: 10.1016/j.molcel.2009.04.023 PMID 19450533
- Mehra, A.; Shi, M.; Baker, C.L.; Colot, H.V.; Loros, J.J.; Dunlap, J.C. (2009). "A role for casein kinase 2 in the mechanism underlying circadian temperature compensation." Cell. 137(4): 749-60. doi: 10.1016/j.cell.2009.03.019 PMID 19450520
- Larrondo, L.F.; Olivares-Yañez, C.; Baker, C.L.; Loros, J.J.; Dunlap, J.C. (2015). "Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination." Science. 347(6221): 1257277. doi: 10.1126/science.1257277 PMID 25635104
Books
- Dunlap, J. C., Loros, J. J., & DeCoursey, P. J. (2004). Chronobiology: Biological timekeeping. Sinauer Associates. ISBN 978-0-87893-396-9
Other works
- A 2015 NPR article, "Why Some Mushrooms Glow In The Dark", notes work done in Dunlap's lab identifying circadian control of bioluminescence in mushrooms.[30]
References
- ↑ 1.0 1.1 1.2 1.3 Gabrielsen, Paul (2015-08-24). "Profile of Jay C. Dunlap" (in en). Proceedings of the National Academy of Sciences 112 (38): 11745–11747. doi:10.1073/pnas.1514590112. ISSN 0027-8424. PMID 26305970. Bibcode: 2015PNAS..11211745G.
- ↑ 2.0 2.1 2.2 Bell-Pedersen D; Borkovich K (2009). "The 2009 George W. Beadle Award". Genetics 551: 29–30.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Dunlap J (2008). "Salad Days in the Rhythms Trade". Genetics 178 (1): 1–13. doi:10.1534/genetics.104.86496. PMID 18202353.
- ↑ 4.0 4.1 "Jay C. Dunlap, PhD – Faculty Expertise Database – Geisel School of Medicine at Dartmouth" (in en-US). https://geiselmed.dartmouth.edu/faculty/facultydb/view.php/?uid=125.
- ↑ Hastings, Michael (1998-12-19). "The brain, circadian rhythms, and clock genes". BMJ: British Medical Journal 317 (7174): 1704–1707. doi:10.1136/bmj.317.7174.1704. ISSN 0959-8138. PMID 9857134.
- ↑ Mirsky, Henry P.; Liu, Andrew C.; Welsh, David K.; Kay, Steve A.; Doyle, Francis J. (2009-07-07). "A model of the cell-autonomous mammalian circadian clock" (in en). Proceedings of the National Academy of Sciences 106 (27): 11107–11112. doi:10.1073/pnas.0904837106. ISSN 0027-8424. PMID 19549830. Bibcode: 2009PNAS..10611107M.
- ↑ 7.0 7.1 7.2 Gooch, V.D.; Mehra, A; Larrondo, L.F.; Fox, J; Touroutoutoudis, M; Loros, J.J.; Dunlap, J.C. (2008-01-01). "Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock.". Eukaryot Cell 7 (1): 28–37. doi:10.1128/EC.00257-07. PMID 17766461.
- ↑ 8.0 8.1 Jay C Dunlap (1999). "Molecular Bases for Circadian Clocks". Cell 96 (2): 271–290. doi:10.1177/0748730411401579. PMID 9988221.
- ↑ Aronson, B. D.; Johnson, K. A.; Loros, J. J.; Dunlap, J. C. (1994-03-18). "Negative feedback defining a circadian clock: autoregulation of the clock gene frequency" (in en). Science 263 (5153): 1578–1584. doi:10.1126/science.8128244. ISSN 0036-8075. PMID 8128244. Bibcode: 1994Sci...263.1578A. https://www.science.org/doi/10.1126/science.8128244.
- ↑ Dunlap, J. C.; Loros, J. J.; Colot, H. V.; Mehra, A.; Belden, W. J.; Shi, M.; Hong, C. I.; Larrondo, L. F. et al. (2007-01-01). "A circadian clock in Neurospora: how genes and proteins cooperate to produce a sustained, entrainable, and compensated biological oscillator with a period of about a day". Cold Spring Harbor Symposia on Quantitative Biology 72: 57–68. doi:10.1101/sqb.2007.72.072. ISSN 0091-7451. PMID 18522516.
- ↑ Loros, J. J.; Dunlap, J. C. (2001-01-01). "Genetic and molecular analysis of circadian rhythms in Neurospora". Annual Review of Physiology 63: 757–794. doi:10.1146/annurev.physiol.63.1.757. ISSN 0066-4278. PMID 11181975. https://www.annualreviews.org/doi/full/10.1146/annurev.physiol.63.1.757.
- ↑ Baker, Christopher L.; Kettenbach, Arminja N.; Loros, Jennifer J.; Gerber, Scott A.; Dunlap, Jay C. (2009-05-15). "Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock". Molecular Cell 34 (3): 354–363. doi:10.1016/j.molcel.2009.04.023. ISSN 1097-4164. PMID 19450533.
- ↑ Mehra, Arun; Shi, Mi; Baker, Christopher L.; Colot, Hildur V.; Loros, Jennifer J.; Dunlap, Jay C. (2009-05-15). "A role for Casein Kinase 2 in the mechanism underlying circadian temperature compensation". Cell 137 (4): 749–760. doi:10.1016/j.cell.2009.03.019. ISSN 0092-8674. PMID 19450520.
- ↑ Hurley, Jennifer M.; Larrondo, Luis F.; Loros, Jennifer J.; Dunlap, Jay C. (2013-12-26). "Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ". Molecular Cell 52 (6): 832–843. doi:10.1016/j.molcel.2013.11.005. ISSN 1097-4164. PMID 24316221.
- ↑ 15.0 15.1 Dunlap, Jay C.; Loros, Jennifer J. (2017-05-19). "Making Time: Conservation of Biological Clocks from Fungi to Animals" (in en). Microbiology Spectrum 5 (3). doi:10.1128/microbiolspec.FUNK-0039-2016. ISSN 2165-0497. PMID 28527179.
- ↑ Crosthwaite, S. K.; Loros, J. J.; Dunlap, J. C. (1995-06-30). "Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript". Cell 81 (7): 1003–1012. doi:10.1016/s0092-8674(05)80005-4. ISSN 0092-8674. PMID 7600569.
- ↑ Shigeyoshi, Y.; Taguchi, K.; Yamamoto, S.; Takekida, S.; Yan, L.; Tei, H.; Moriya, T.; Shibata, S. et al. (1997-12-26). "Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript". Cell 91 (7): 1043–1053. doi:10.1016/s0092-8674(00)80494-8. ISSN 0092-8674. PMID 9428526.
- ↑ 18.0 18.1 Crosthwaite, S.K.; Dunlap, J.C.; Loros, J.J. (1997-05-02). "Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity.". Science 276 (5313): 263–269. doi:10.1126/science.276.5313.763. ISSN 0036-8075. PMID 9115195. https://www.science.org/doi/10.1126/science.276.5313.763.
- ↑ Dunlap, Jay (1998-06-05). "An End in the Beginning" (in en). Science 280 (5369): 1548–1549. doi:10.1126/science.280.5369.1548. ISSN 0036-8075. PMID 9644021. https://www.science.org/doi/10.1126/science.280.5369.1548.
- ↑ Tseng, Yu-Yao; Hunt, Suzanne M.; Heintzen, Christian; Crosthwaite, Susan K.; Schwartz, Jean-Marc (2012-03-29). "Comprehensive Modelling of the Neurospora Circadian Clock and Its Temperature Compensation" (in en). PLOS Computational Biology 8 (3): e1002437. doi:10.1371/journal.pcbi.1002437. ISSN 1553-7358. PMID 22496627. Bibcode: 2012PLSCB...8E2437T.
- ↑ Froehlich, Allan C.; Liu, Yi; Loros, Jennifer J.; Dunlap, Jay C. (2002-08-02). "White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter". Science 297 (5582): 815–819. doi:10.1126/science.1073681. ISSN 1095-9203. PMID 12098706. Bibcode: 2002Sci...297..815F.
- ↑ Dunlap, Jay C.; Loros, Jennifer J. (2006-10-24). "How fungi keep time: circadian system in Neurospora and other fungi". Current Opinion in Microbiology 9 (6): 579–587. doi:10.1016/j.mib.2006.10.008. ISSN 1369-5274. PMID 17064954. https://pubmed.ncbi.nlm.nih.gov/17064954/.
- ↑ Loros, J. J.; Denome, S. A.; Dunlap, J. C. (1989-01-20). "Molecular cloning of genes under control of the circadian clock in Neurospora". Science 243 (4889): 385–388. doi:10.1126/science.2563175. ISSN 0036-8075. PMID 2563175. Bibcode: 1989Sci...243..385L. https://pubmed.ncbi.nlm.nih.gov/2563175/.
- ↑ Loros, J. J.; Dunlap, J. C.; Larrondo, L. F.; Shi, M.; Belden, W. J.; Gooch, V. D.; Chen, C.-H.; Baker, C. L. et al. (2007-01-01). "Circadian output, input, and intracellular oscillators: insights into the circadian systems of single cells". Cold Spring Harbor Symposia on Quantitative Biology 72: 201–214. doi:10.1101/sqb.2007.72.067. ISSN 0091-7451. PMID 18419278.
- ↑ Pregueiro, António M.; Liu, Qiuyun; Baker, Christopher L.; Dunlap, Jay C.; Loros, Jennifer J. (2006-08-04). "The Neurospora Checkpoint Kinase 2: A Regulatory Link Between the Circadian and Cell Cycles" (in en). Science 313 (5787): 644–649. doi:10.1126/science.1121716. ISSN 0036-8075. PMID 16809488. Bibcode: 2006Sci...313..644P. https://www.science.org/doi/10.1126/science.1121716.
- ↑ Hurley, Jennifer M.; Jankowski, Meaghan S.; De Los Santos, Hannah; Crowell, Alexander M.; Fordyce, Samuel B.; Zucker, Jeremy D.; Kumar, Neeraj; Purvine, Samuel O. et al. (2018-12-26). "Circadian Proteomic Analysis Uncovers Mechanisms of Post-Transcriptional Regulation in Metabolic Pathways". Cell Systems 7 (6): 613–626.e5. doi:10.1016/j.cels.2018.10.014. ISSN 2405-4712. PMID 30553726.
- ↑ Shi, Mi; Larrondo, Luis F.; Loros, Jennifer J.; Dunlap, Jay C. (2007-12-11). "A developmental cycle masks output from the circadian oscillator under conditions of choline deficiency in Neurospora". Proceedings of the National Academy of Sciences of the United States of America 104 (50): 20102–20107. doi:10.1073/pnas.0706631104. ISSN 1091-6490. PMID 18056807. Bibcode: 2007PNAS..10420102S.
- ↑ Larrondo, Luis F.; Olivares-Yañez, Consuelo; Baker, Christopher L.; Loros, Jennifer J.; Dunlap, Jay C. (2015-01-30). "Decoupling circadian clock protein turnover from circadian period determination". Science 347 (6221): 1257277. doi:10.1126/science.1257277. ISSN 1095-9203. PMID 25635104.
- ↑ Oliveira, Anderson G.; Stevani, Cassius V.; Waldenmaier, Hans E.; Viviani, Vadim; Emerson, Jillian M.; Loros, Jennifer J.; Dunlap, Jay C. (2015-03-30). "Circadian Control Sheds Light on Fungal Bioluminescence" (in English). Current Biology 25 (7): 964–968. doi:10.1016/j.cub.2015.02.021. ISSN 0960-9822. PMID 25802150.
- ↑ 30.0 30.1 Waldenmaier, Hans; Chemist, Research; Paulo, University of Sao. "Why Some Mushrooms Glow In The Dark". https://www.npr.org/2015/03/21/394089178/why-some-mushrooms-glow-in-the-dark.
- ↑ Fuller, Kevin K.; Cramer, Robert A.; Zegans, Michael E.; Dunlap, Jay C.; Loros, Jennifer J. (2016-09-20). "Aspergillus fumigatus Photobiology Illuminates the Marked Heterogeneity between Isolates". mBio 7 (5). doi:10.1128/mBio.01517-16. ISSN 2150-7511. PMID 27651362.
- ↑ "Neurospora crassa genome project | Broad Institute" (in en). 2016-09-14. https://www.broadinstitute.org/fungal-genome-initiative/neurospora-crassa-genome-project.
- ↑ Hurley, Jennifer M.; Loros, Jennifer J.; Dunlap, Jay C. (2016-05-01). "The circadian system as an organizer of metabolism". Fungal Genetics and Biology 90: 39–43. doi:10.1016/j.fgb.2015.10.002. PMID 26498192.
- ↑ Dekhang, Rigzin; Wu, Cheng; Smith, Kristina M.; Lamb, Teresa M.; Peterson, Matthew; Bredeweg, Erin L.; Ibarra, Oneida; Emerson, Jillian M. et al. (2016-11-15). "The Neurospora Transcription Factor ADV-1 Transduces Light Signals and Temporal Information to Control Rhythmic Expression of Genes Involved in Cell Fusion". G3: Genes, Genomes, Genetics 7 (1): 129–142. doi:10.1534/g3.116.034298. ISSN 2160-1836. PMID 27856696.
- ↑ Pelham, Jacqueline F.; Dunlap, Jay C.; Hurley, Jennifer M. (2020-11-11). "Intrinsic disorder is an essential characteristic of components in the conserved circadian circuit". Cell Communication and Signaling 18 (1): 181. doi:10.1186/s12964-020-00658-y. ISSN 1478-811X. PMID 33176800.
- ↑ Kelliher, C.M.; Lambreghts, R.; Xiang, Q.; Baker, C.L.; Loros, J.J.; Dunlap, J.C. (2020-12-09). "PRD-2 directly regulates casein kinase I and counteracts nonsense-mediated decay in the Neurospora circadian clock.". eLife 9. doi:10.7554/eLife.64007. PMID 33295874.
 KSF
KSF