Dynamin
Topic: Biology
 From HandWiki - Reading time: 6 min
From HandWiki - Reading time: 6 min
| Dynamin family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin I from Rattus norvegicus | |||||||||
| Identifiers | |||||||||
| Symbol | Dynamin_N | ||||||||
| Pfam | PF00350 | ||||||||
| Pfam clan | CL0023 | ||||||||
| InterPro | IPR001401 | ||||||||
| PROSITE | PDOC00362 | ||||||||
| |||||||||
| Dynamin central region | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin I from Rattus norvegicus | |||||||||
| Identifiers | |||||||||
| Symbol | Dynamin_M | ||||||||
| Pfam | PF01031 | ||||||||
| InterPro | IPR000375 | ||||||||
| |||||||||
Dynamin is a GTPase responsible for endocytosis in the eukaryotic cell. Dynamin is part of the "dynamin superfamily", which includes classical dynamins, dynamin-like proteins, Mx proteins, OPA1, mitofusins, and GBPs. Members of the dynamin family are principally involved in the scission of newly formed vesicles from the membrane of one cellular compartment and their targeting to, and fusion with, another compartment, both at the cell surface (particularly caveolae internalization) as well as at the Golgi apparatus.[1][2][3] Dynamin family members also play a role in many processes including division of organelles,[4] cytokinesis and microbial pathogen resistance.
Structure
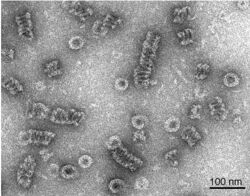
Dynamin itself is a 96 kDa enzyme, and was first isolated when researchers were attempting to isolate new microtubule-based motors from the bovine brain. Dynamin has been extensively studied in the context of clathrin-coated vesicle budding from the cell membrane.[3][6] Beginning from the N-terminus, Dynamin consists of a GTPase domain connected to a helical stalk domain via a flexible neck region containing a Bundle Signalling Element and GTPase Effector Domain. At the opposite end of the stalk domain is a loop that links to a membrane-binding Pleckstrin homology domain. The protein strand then loops back towards the GTPase domain and terminates with a Proline Rich Domain that binds to the Src Homology domains of many proteins.
Function
During clathrin-mediated endocytosis, the cell membrane invaginates to form a budding vesicle. Dynamin binds to and assembles around the neck of the endocytic vesicle, forming a helical polymer arranged such that the GTPase domains dimerize in an asymmetric manner across helical rungs.[7][8] The polymer constricts the underlying membrane upon GTP binding and hydrolysis via conformational changes emanating from the flexible neck region that alters the overall helical symmetry.[8] Constriction around the vesicle neck leads to the formation of a hemi-fission membrane state that ultimately results in membrane scission.[2][6][9] Constriction may be in part the result of the twisting activity of dynamin, which makes dynamin the only molecular motor known to have a twisting activity.[10]
Types
In mammals, three different dynamin genes have been identified with key sequence differences in their Pleckstrin homology domains leading to differences in the recognition of lipid membranes:
- Dynamin I is expressed in neurons and neuroendocrine cells
- Dynamin II is expressed in most cell types
- Dynamin III is strongly expressed in the testis, but is also present in heart, brain, and lung tissue.[1][6]
Pharmacology
Small molecule inhibitors of dynamin activity have been developed, including Dynasore[11][12] and photoswitchable derivatives (Dynazo)[13] for spatiotemporal control of endocytosis with light (photopharmacology).
Disease implications
Mutations in Dynamin II have been found to cause dominant intermediate Charcot-Marie-Tooth disease.[14] Epileptic encephalopathy–causing de novo mutations in dynamin have been suggested to cause dysfunction of vesicle scission during synaptic vesicle endocytosis.[15]
References
- ↑ 1.0 1.1 Henley, John R.; Cao, Hong; McNiven, Mark A. (December 16, 1999). "Participation of dynamin in the biogenesis of cytoplasmic vesicles". The FASEB Journal 13 (9002): S243-7. doi:10.1096/fasebj.13.9002.S243. PMID 10619136. https://onlinelibrary.wiley.com/doi/abs/10.1096/fasebj.13.9002.S243.
- ↑ 2.0 2.1 Hinshaw, J. "Research statement, Jenny E. Hinshaw, Ph.D." National Institute of Diabetes & Digestive & Kidney Diseases, Laboratory of Cell Biochemistry and Biology. Accessed 19 March 2013.
- ↑ 3.0 3.1 "The dynamins: redundant or distinct functions for an expanding family of related GTPases?". Proceedings of the National Academy of Sciences of the United States of America 94 (2): 377–384. January 1997. doi:10.1073/pnas.94.2.377. PMID 9012790. Bibcode: 1997PNAS...94..377U.
- ↑ "Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation". The FEBS Journal 272 (20): 5169–5181. October 2005. doi:10.1111/j.1742-4658.2005.04939.x. PMID 16218949.
- ↑ "Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding". Nature 374 (6518): 190–192. March 1995. doi:10.1038/374190a0. PMID 7877694. Bibcode: 1995Natur.374..190H.
- ↑ 6.0 6.1 6.2 "The dynamin superfamily: universal membrane tubulation and fission molecules?". Nature Reviews. Molecular Cell Biology 5 (2): 133–147. February 2004. doi:10.1038/nrm1313. PMID 15040446.
- ↑ "A dynamin mutant defines a superconstricted prefission state". Cell Reports 8 (3): 734–742. August 2014. doi:10.1016/j.celrep.2014.06.054. PMID 25088425.
- ↑ 8.0 8.1 "Cryo-EM of the dynamin polymer assembled on lipid membrane". Nature 560 (7717): 258–262. August 2018. doi:10.1038/s41586-018-0378-6. PMID 30069048. Bibcode: 2018Natur.560..258K.
- ↑ "A hemi-fission intermediate links two mechanistically distinct stages of membrane fission". Nature 524 (7563): 109–113. August 2015. doi:10.1038/nature14509. PMID 26123023. Bibcode: 2015Natur.524..109M.
- ↑ "GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission". Nature 441 (7092): 528–531. May 2006. doi:10.1038/nature04718. PMID 16648839. Bibcode: 2006Natur.441..528R.
- ↑ "Dynasore, a cell-permeable inhibitor of dynamin" (in English). Developmental Cell 10 (6): 839–850. June 2006. doi:10.1016/j.devcel.2006.04.002. PMID 16740485.
- ↑ "Modulation of dynamin function by small molecules". Biological Chemistry 399 (12): 1421–1432. November 2018. doi:10.1515/hsz-2018-0257. PMID 30067507.
- ↑ "Correction: Photoswitchable dynasore analogs to control endocytosis with light". Chemical Science 11 (35): 9712. September 2020. doi:10.1039/D0SC90189J. PMID 33016959.
- ↑ "Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease". Nature Genetics 37 (3): 289–294. March 2005. doi:10.1038/ng1514. PMID 15731758.
- ↑ "Epileptic encephalopathy-causing mutations in DNM1 impair synaptic vesicle endocytosis". Neurology. Genetics 1 (1): e4. June 2015. doi:10.1212/01.NXG.0000464295.65736.da. PMID 27066543.
External links
- Dynamins at the US National Library of Medicine Medical Subject Headings (MeSH)
 |
 KSF
KSF