Glossary of evolutionary biology
Topic: Biology
 From HandWiki - Reading time: 54 min
From HandWiki - Reading time: 54 min
Short description: List of definitions of terms and concepts commonly used in evolutionary biology
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.
A
- adaptation
- 1. The dynamic evolutionary process by which biological organisms develop characteristics that allow them to survive and reproduce within their environments.
- 2. The state or condition reached by a population during that process.
- 3. Any character or phenotypic trait with a functional role in an individual organism and which has evolved and is maintained through natural selection.
- adaptationism
- The Darwinian view that many or most physiological and behavioral traits of organisms are adaptations that have evolved for specific functions or for specific reasons (as opposed to being byproducts of the evolution of other traits, consequences of biological constraints, or the result of random variation).
- adaptive radiation
- The simultaneous or near-simultaneous evolutionary divergence of multiple members of a single phylogenetic lineage into a variety of different forms with different adaptations, especially a diversification in the use of resources or habitats.[1]
- agamospecies
- A species that does not reproduce sexually but rather by cloning.[2] Agamospecies are sometimes represented by species complexes that contain some diploid individuals and other apomictic forms—in particular, plant species that can reproduce via agamospermy.[3]
- allele
- One of multiple alternative versions of an individual gene, each of which is a viable DNA sequence occupying a given position, or locus, on a chromosome. For example, in humans, one allele of the eye-color gene produces blue eyes and another allele of the eye-color gene produces brown eyes.
- allele frequency
- The relative frequency with which a particular allele of a given gene (as opposed to other alleles of the same gene) occurs at a particular locus in the members of a population; more specifically, it is the proportion of all chromosomes within a population that carry a particular allele, expressed as a fraction or percentage. Allele frequency is distinct from genotype frequency, although they are related.
- allochronic isolation
- The isolation of two populations of a species due to a change in breeding periods. This isolation acts as a precursor to allochronic speciation, a type of speciation which results when two populations of a species become isolated due to differences in reproductive timing. An example is the periodical 13- and 17-year Magicicada species.[3]
- allo-parapatric speciation
- A mode of speciation where divergence occurs in allopatry and is completed upon secondary contact of the populations--effectively a form of reinforcement.[4][3]
- allometry
- The comparative study of the relationship between the size of an organism's body (or of a specific organ, e.g. the brain) and various other biological characteristics, such as body shape, anatomy, physiology, or behavior.
- allopatric speciation
- A mode of speciation where the evolution of reproductive isolation is caused by the geographic separation of two or more populations of a single species.[5]
- allopatric taxa
- Specific species that are allopatrically distributed.
- allopatry
- The phenomenon by which two or more populations of a single species exist in geographic isolation from one another.
- allopolyploid
- A polyploid cell or organism in which the several sets of chromosomes originate from more than one species, as in an intraspecific hybrid.[1]
- allo-sympatric speciation
- A mode of speciation where divergence occurs in allopatry and is completed upon secondary contact of the populations–effectively a form of reinforcement.[6][3]
- altruism
- anagenesis
- Evolutionary change that occurs within a species lineage as opposed to lineage splitting (cladogenesis).[7]
- analogous structures
- A set of morphological structures in different organisms which have similar form or function but were not present in the organisms' last common ancestor. The cladistic term for the same phenomenon is homoplasy.
- ancestor
- ancestral trait
- For a given clade, any trait or feature (e.g. a specific phenotype) that appears in the clade's common ancestor; the same trait may also appear in some or all of the lineal descendants included within the clade, indicating that it has undergone little or no significant change during the clade's evolutionary history and thus retained its "primitive" condition. Some but not all subgroups within the clade may contain derived traits, in which the ancestral trait has changed significantly over evolutionary time such that the original ancestral condition no longer exists. Both terms are relative: an ancestral trait for one clade may be a derived trait for a different clade. The term "ancestral trait" is often used interchangeably with the more technical term plesiomorphy.
- anticipation
- A phenomenon by which the symptoms of a genetic disorder become apparent (and often more severe) at an earlier age in affected individuals with each generation that inherits the disorder.
- apomorphy
- A derived character state; i.e. the state or condition of a particular trait or feature (e.g. a specific phenotype) that is distinct from and derivative of an ancestral character by virtue of its modification over time in one or more lineal descendants of a given clade. Apomorphies are often viewed as evolutionary "innovations" which set the taxa in which they appear apart from the clade's common ancestor, as well as from other clades; shared apomorphies are used to construct and define clades. The term is relative; a trait considered an apomorphy in one clade may not be considered an apomorphy in a different clade. Contrast plesiomorphy.
- aptation
- Any character or phenotypic trait that is currently subject to natural selection, whether its origin can be ascribed to selective processes (adaptation) or to processes other than selection or selection for a function that is different from the current function (exaptation).[8]
- area cladogram
- asexual reproduction
- associative overdominance
- The phenomenon by which the linkage of a neutral locus to a selectively maintained polymorphism causes the heterozygosity of the neutral locus to increase.[8]
- assortative mating
- A mating system in which individuals with similar phenotypes mate with each other more frequently than would be expected in a completely random mating system. Assortative mating usually has the effect of increasing genetic relatedness between members of the mating population. Contrast disassortative mating.
- atavism
- A modification of a biological structure whereby an ancestral trait suddenly reappears after having been lost through evolutionary change in previous generations.[9] Atavisms can occur in a number of different ways, including by the re-expression of latent genes for ancestral phenotypes as a result of mutation, or by the shortening or prolongation of the time allocated for the ontogenesis of a particular trait during development.
- autapomorphy
- autoallopolyploid
- autopolyploid
- autozygote
- A cell or organism that is homozygous for a locus at which the two homologous alleles are identical by descent, both having been derived from a single gene in a common ancestor.[8] Contrast allozygote.

In allopatric speciation, a population becomes separated by a geographic barrier and reproductive isolation results in two separate species.
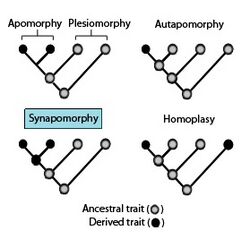
An apomorphy is a derived trait present in one or more members of a clade but not the common ancestor; a plesiomorphy is an ancestral trait present in the common ancestor of the clade and possibly some or all of its descendants.
B
- backcrossing
- The breeding of a hybrid organism with one of its parents or an individual genetically similar to one of its parents, often intentionally as a type of selective breeding, with the aim of producing offspring with a genetic identity which is closer to that of the parent. The reproductive event and the resulting progeny are both referred to as a backcross, often abbreviated in genetics shorthand with the symbol BC.
- Bateson–Dobzhansky–Muller model
- An evolutionary model of the genetic incompatibility that occurs as a result of negative epistatic interactions between two or more genes or alleles with different evolutionary histories, which may meet when distinct populations hybridize. The incompatible genes or alleles themselves, referred to as Dobzhansky–Muller incompatibilities, may be the result of random or neutral mutations, or they may be specific adaptations driven by natural selection. By preventing populations from successfully interbreeding, these incompatibilities can reinforce reproductive isolation and thereby increase the chance of speciation.
- behavioral isolation
- biogeography
- The scientific study of the spatial distributions of biological organisms, populations, and species. It includes the study of both extinct and extant organisms.[10]
- biological constraints
- biological species concept
- bottleneck
- See population bottleneck.
C
- canalisation
- The ability of a population to consistently produce the same phenotype regardless of the variability of its environment or the genetic variation within its genome. The concept is most often used in developmental biology to interpret the observation that developmental pathways are frequently shaped by natural selection such that developing cell lineages are "guided" or "canalized" towards a single, definite fate, becoming progressively more resistant to any minor perturbations that may redirect development of the cells away from their initial course.
- carrier
- An individual who has inherited a recessive allele for a genetic trait or mutation but in whom the trait is not usually expressed or observable in the phenotype. Carriers are usually heterozygous for the recessive allele and therefore still able to pass the allele onto their offspring, where the associated phenotype may reappear if the offspring inherits another copy of the allele. The term is commonly used in medical genetics in the context of a disease-causing recessive allele.
- centrifugal speciation
- A variation of peripatric speciation in which speciation occurs by geographic isolation, but reproductive isolation evolves in the larger population instead of the peripherally isolated population.[11]
- character displacement
- The phenomenon by which differences between similar species that occupy similar niches and have partially overlapping geographic distributions are accentuated in regions where the species co-occur but are minimized or lost where the species' distributions do not overlap. This occurs because competition between the similar species for one or more limited resources drives evolutionary change that differentiates the species in the common geographic areas such that they no longer occupy the same niche, thereby allowing them to coexist and avoiding competitive exclusion.
- chimerism
- The presence of two or more populations of cells with distinct genotypes within the body of an individual organism, known as a chimera, which has developed from the fusion of cells originating from separate zygotes; each population of cells retains its own genome, such that the organism as a whole is a mixture of genetically non-identical tissues. Genetic chimerism may be inherited (e.g. by the fusion of multiple embryos during pregnancy) or acquired after birth (e.g. by allogeneic transplantation of cells, tissues, or organs from a genetically non-identical donor); in plants, it can result from grafting or errors in cell division. It is similar to but distinct from mosaicism.
- chromosomal speciation
- chronospecies
- clade
- A phylogenetic grouping of organisms that consists of a single common ancestor and all of its lineal descendants, and which by definition is monophyletic. The common ancestor may be an individual organism, a population, a species, or any other taxon; any and all members of a clade may be extant or extinct. Clades can be visualized with cladograms and are the basis of cladistics.
- cladistics
- An approach to biological classification in which organisms are grouped in clades defined by shared ancestry; hypothesized relationships between organisms are typically based on shared derived characters which can be traced to the most recent common ancestor and are not present in more distant ancestors or unrelated groups.
- cladogenesis
- The splitting of a single species lineage within a phylogeny into multiple lineages.[7]
- cladogram
- classical genetics
- The branch of genetics based solely on observation of the visible results of reproductive acts, as opposed to that made possible by the modern techniques and methodologies of molecular biology. Contrast molecular genetics.
- cline
- A measurable spatial gradient in a single biological character or trait of a species or population across its geographic range. The nature of a cline may be genotypic (e.g. variation in allele frequency) or phenotypic (e.g. variation in body size or pigmentation), and may show smooth, continuous gradation or abrupt changes between different geographic regions.
- cloning
- The process of producing, either naturally or artificially, individual organisms or cells which are genetically identical to each other. Clones are the result of all forms of asexual reproduction, and cells that undergo mitosis produce daughter cells that are clones of the parent cell and of each other. Cloning may also refer to biotechnology methods which artificially create copies of organisms or cells, or, in molecular cloning, copies of DNA fragments or other molecules.
- cluster analysis
- clustering
- co-operation
- coadaptation
- The mutual adaptation of organisms belonging to different populations or species, of different parts of the same organism, or of genes at different loci in the same genome, especially implying that adaptation in both entities is driven by the same evolutionary force.[12]
- codominance
- coevolution
- The process by which two or more distinct populations, species, or other groups of organisms, or two or more distinct traits within a species, reciprocally affect each other's evolution through natural selection. Each party in a coevolutionary relationship exerts selective pressures upon the other, leading to the evolution of separate traits in each party.
- cohesion species concept
- colonization
- The spread of a population to a new geographic area.
- common ancestor
- An organism or taxon (e.g. a species) which is hypothesized to be the lineal progenitor of two or more organisms or taxa which exist at a later point in evolutionary time. The concept of common descent is fundamental to the study of evolution, phylogenetics, and cladistics; for instance, all clades, by definition, are rooted in a common ancestor. See also most recent common ancestor.
- competitive gametic isolation
- complex trait
- See quantitative trait.
- congruent clines
- consanguineous
- (of two or more individuals) Closely genetically related; sharing a recent common ancestor (usually no more than three or four generations distant). The effect of consanguineous mating, also known as inbreeding, is to increase the probability that the progeny will be homozygous at any given pair of genetic loci.[13]
- conservation genetics
- An interdisciplinary branch of population genetics which applies genetic methods and concepts in an effort to understand the dynamics of genes in populations, with a principal aim of avoiding extinctions and preserving and restoring biodiversity.
- convergent evolution
- The independent evolution of similar traits or adaptations in two or more different taxa from different periods or epochs in time, creating analogous structures that have similar form or function but were not present in the last common ancestor of those taxa; e.g. structures enabling flight evolved independently in at least four distinct lineages: insects, birds, pterosaurs, and bats. In cladistics, the same phenomenon is termed homoplasy. Contrast divergent evolution.
- copulatory behavioral isolation
- coupling
- court jester hypothesis
- cospeciation
- A type of speciation in which more than two species speciate concurrently due to their ecological associations (e.g. host-parasite interactions).[14]
- crossbreeding
- The breeding of purebred parents belonging to two different breeds, varieties, or populations, often intentionally as a type of selective breeding, with the aim of producing offspring which share traits of both parent lineages or which show heterosis. In animal breeding, the progeny of a cross between breeds of the same species is called a crossbreed, whereas the progeny of a cross between different species is called a hybrid.
- crown group
- cryptic species
- cytoplasmic isolation
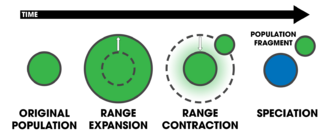
In centrifugal speciation, the range of an original population (green) expands and then contracts, leaving an isolated fragment population behind. In the absence of interbreeding, the central population (changed to blue) becomes reproductively isolated over time.
D
- Darwinism
- The understanding of biological evolution as developed by the English naturalist Charles Darwin and others, which states that all biological organisms arise and develop through the natural selection of small, inherited variations that increase the individual's ability to compete, survive, and reproduce. Colloquially, the term is sometimes used to refer more broadly to modern evolutionary theory as a whole, though in scientific circles distinctions are usually made between Darwin's ideas and later additions to evolutionary biology.
- de novo mutation
- A spontaneous mutation in the genome of an individual organism that is new to that organism's lineage, having first appeared in a germ cell of one of the organism's parents or in the fertilized egg that develops into the organism; i.e. a mutation that was not present in either parent's genome.[8]
- de-extinction
- derived trait
- For a given clade, any trait or feature (e.g. a specific phenotype) that is present within one or more subgroups of the clade but not in the clade's common ancestor. Derived traits show significant differences from the original "primitive" condition of an ancestral trait found in the common ancestor, implying that the trait has undergone extensive adaptation during the clade's evolutionary history to reach its derivative condition. Both terms are relative: a derived trait for one clade may be an ancestral trait for a different clade. The term "derived trait" is often used interchangeably with the more technical term apomorphy.
- descendant
- developmental biology
- diploid
- (of a cell or organism) Having two homologous copies of each chromosome. Contrast haploid and polyploid.
- directional selection
- A mode of natural selection in which an extreme trait or phenotype is favored over other phenotypes, causing allele frequencies to shift over time in the direction of that trait or phenotype. This shift can occur whether or not the alleles governing the extreme phenotype are dominant or recessive.
- directional speciation
- disassortative mating
- A mating system in which individuals with dissimilar phenotypes mate with each other more frequently than would be expected in a completely random mating system. Disassortative mating usually has the effect of decreasing genetic relatedness between members of the mating population. Contrast assortative mating.
- dispersal
- disruptive selection
- A mode of natural selection in which the extreme values of a trait or phenotype within a breeding population are favored over intermediate values, causing allele frequencies to shift over time away from the intermediate. This causes the variance in the trait to increase and results in the population dividing into two distinct groups, each with trait values at one end of the trait's distribution curve.
- divergence-with-gene-flow
- divergent evolution
- The process by which any phenotypic or genotypic distinction emerges between two different populations or evolutionary lineages. Divergence may occur by any of a variety of mechanisms but is often especially noticeable after the two lineages have been reproductively isolated for many generations.[7]
- diversification
- Dobzhansky–Muller model
- See Bateson–Dobzhansky–Muller model.
- dominance
- A relationship between the alleles of a gene in which one allele produces an effect on phenotype that overpowers or "masks" the contribution of another allele at the same locus; the first allele and its associated phenotypic trait are said to be dominant, and the second allele and its associated trait are said to be recessive. Often, the dominant allele codes for a functional protein while its recessive counterpart does not. Dominance is not an inherent property of any allele or phenotype, but simply describes its relationship to one or more other alleles or phenotypes; it is possible for one allele to be simultaneously dominant over a second allele, recessive to a third, and codominant to a fourth. In genetics shorthand, dominant alleles are often represented by a single uppercase letter (e.g. "A", in contrast to the recessive "a").
- dosage compensation
- Any mechanism by which organisms neutralize the large difference in gene dosage caused by the presence of differing numbers of sex chromosomes in the different sexes, thereby equalizing the expression of sex-linked genes so that the members of each sex receive the same or similar amounts of the products of such genes. An example is X-inactivation in female mammals.
E
- ecogeographic isolation
- ecological allopatry
- ecological character displacement
- ecological genetics
- The study of genetics as it pertains to the ecology and fitness of natural populations of living organisms.
- ecological isolation
- ecological niche
- See niche.
- ecological speciation
- A type of speciation in which reproductive isolation is caused by the interaction of individuals of a species with their environment.[15]
- ecological species concept
- emergenesis
- The quality of genetic traits that results from a specific configuration of interacting genes, rather than simply their combination.
- endemism
- The ecological state of a species being unique to a single geographic location, such as an island, nation, country, or any other clearly defined area, or to a single habitat type.
- environmental gradient
- epigenetics
- epistasis
- The collective action of multiple genes interacting during gene expression. A form of gene action, epistasis can be either additive or multiplicative in its effects on specific phenotypic traits.
- error catastrophe
- The extinction of a population of organisms (insofar as the population can be defined by one or more identifiable characteristics) as a result of the excessive accumulation of genetic mutations, such that the population loses self-identity because all of its mutated descendants lack the identifiable characteristics.
- ethological isolation
- ethological pollinator isolation
- euploidy
- The condition of a cell or organism having an abnormal number of complete sets of chromosomes, possibly excluding the sex chromosomes. Euploidy differs from aneuploidy, in which a cell or organism has an abnormal number of one or more specific individual chromosomes.
- evolution
- The phenomenon by which the heritable characteristics of biological populations change over successive generations. Evolution occurs when processes such as natural selection and genetic drift act on the variation in characteristics that exists between members of a population, resulting in certain characteristics becoming more or less common within the population.
- evolutionary arms race
- The positive feedback mechanism operating between competing sets of co-evolving genes, traits, species, or other taxa which evolve specific adaptations and counter-adaptations due to each other's presence, which may be seen as analogous with an "arms race".
- evolutionary biology
- The discipline of biology that studies the evolution of biological organisms and the processes by which it operates, including natural selection, adaptation, common descent, and speciation. A core element of the modern synthesis, evolutionary biology integrates concepts from genetics, systematics, ecology, paleontology, developmental biology, and numerous other fields.
- evolutionary landscape
- evolutionary species concept
- exaptation
- expressivity
- For a given genotype associated with a variable non-binary phenotype, the proportion of individuals with that genotype who show or express the phenotype to a specified extent, usually given as a percentage. Because of the many complex interactions that govern gene expression, the same allele may produce a wide variety of possible phenotypes of differing qualities or degrees in different individuals; in such cases, both the phenotype and genotype may be said to show variable expressivity. Expressivity attempts to quantify the range of possible levels of phenotypic variation in a population of individuals expressing the phenotype of interest. Compare penetrance.
- extant
- Currently living or existing; still in existence and not extinct. The term is generally used to refer to the present-day state of existence of a particular taxon (such as a family, genus, species, etc.).
- extended evolutionary synthesis
- extinction
- extrinsic hybrid inviability
- extrinsic postzygotic isolation
F
- fitness
- The reproductive success, or propensity to produce offspring, during the lifetime of an individual
- fixation
- The process by which a single allele for a particular gene with multiple alleles increases in frequency in a given population such that it becomes permanently established as the only allele at that locus within the population's gene pool. How long fixation takes depends on selection pressures and chance fluctuations in allele frequencies.
- floral isolation
- flowering asynchrony
- founder effect
- The loss of genetic variation that occurs when a new, physically isolated population is established by a very small number of individuals who have migrated from a larger population and are not fully representative of the larger population's genetic diversity. As a result, the new population is often distinctively different, both genotypically and phenotypically, from the parent population. Besides migration, population bottlenecks can also result in a type of founder effect; extreme founder effects can lead to speciation.
- founder event
- founder-flush-crash
- founder takes all
- A hypothesis that describes the evolutionary advantages of the first-arriving lineages in a new ecosystem.[16] An example could be when a species becomes reproductively isolated on an island, as in peripatric speciation.
- fugitive species
- A species that only temporarily occupies environments or habitats (either because its members frequently migrate or because its environments frequently change) and so does not persist for many generations at any one site.[1]
G
- gametic isolation
- gene
- Any segment or set of segments of a nucleic acid molecule that contains the information necessary to produce a functional RNA transcript in a controlled manner. Genes are often considered the fundamental units of heredity and are typically encoded in DNA. A particular gene can have multiple different versions, or alleles, and a single gene may influence many different phenotypes.
- gene dosage
- The number of copies of a particular gene present in a genome. Gene dosage directly influences the amount of gene product a cell is able to express, though a variety of controls have evolved which tightly regulate gene expression. Changes in gene dosage caused by mutations include copy-number variations.
- gene drive
- gene duplication
- A type of mutation defined as any duplication of a region of DNA that contains a gene. Compare chromosomal duplication.
- gene expression
- The set of processes by which the information encoded in a gene is used in the synthesis of a gene product, such as a protein or non-coding RNA, or otherwise made available to influence one or more phenotypes. Expression at the molecular level canonically consists of transcription and translation, though the information contained within a DNA sequence need not necessarily be transcribed and translated to exert an influence on molecular events, however; broader definitions encompass a huge variety of other ways in which genetic information can be expressed.
- gene flow
- The transfer of genetic variation from one population to another, by any available means, e.g. by sexual reproduction, horizontal gene transfer, or retroviral integration.
- gene pool
- The sum of all of the various alleles shared by the members of a single population.
- gene product
- Any of the biochemical material resulting from the expression of a gene, most commonly interpreted as the functional mRNA transcript produced by transcription of the gene or the fully constructed protein produced by translation of the transcript, though non-coding RNA molecules such as transfer RNAs may also be considered gene products.
- genealogical species concept
- generation
- 1. In any given organism, a single reproductive cycle, or the phase between two consecutive reproductive events, i.e. between an individual organism's reproduction and that of the progeny of that reproduction; or the actual or average length of time required to complete a single reproductive cycle, either for a particular lineage or for a population or species as a whole.
- 2. In a given population, those individuals (often but not necessarily living contemporaneously) who are equally removed from a given common ancestor by virtue of the same number of reproductive events having occurred between them and the ancestor.[13]
- genetic association
- The co-occurrence within a population of one or more alleles or genotypes with a particular phenotypic trait more often than might be expected by chance alone; such statistical correlation may be used to infer that the alleles or genotypes are responsible for producing the given phenotype.
- genetic bottleneck
- See population bottleneck.
- genetic counseling
- The process of advising individuals or families who are affected by or at risk of developing genetic disorders in order to help them understand and adapt to the physiological, psychological, and familial implications of genetic contributions to disease. Genetic counseling integrates genetic testing, genetic genealogy, and genetic epidemiology.[17]
- genetic disorder
- Any illness, disease, or other health problem directly caused by one or more abnormalities in an organism's genome which are congenital (present at birth) and not acquired later in life. Causes may include a mutation to one or more genes, or a chromosomal abnormality such as an aneuploidy of a particular chromosome. The mutation responsible may occur spontaneously during embryonic development or may be inherited from one or both parents, in which case the genetic disorder is also classified as a hereditary disorder. Though the abnormality itself is present before birth, the actual disease it causes may not develop until much later in life; some genetic disorders do not necessarily guarantee eventual disease but simply increase the risk of the organism developing it.
- genetic distance
- A measure of the genetic divergence between species, populations within a species, or individuals, used especially in phylogenetics to express either the time elapsed since the existence of a common ancestor or the degree of differentiation in the DNA sequences comprising the genomes of each population or individual.
- genetic diversity
- The total number of genetic traits or characteristics in the genetic make-up of a population, species, or other group of organisms. It is often used as a measure of the adaptability of a group to changing environments. Genetic diversity is similar to, though distinct from, genetic variability.
- genetic drift
- A change in the frequency with which an existing allele occurs in a population due to random variation in the distribution of alleles from one generation to the next. It is often interpreted as the role that random chance plays in determining whether a given allele becomes more or less common with each generation, irrespective of the influence of natural selection. Genetic drift may cause certain alleles, even otherwise advantageous ones, to disappear completely from the gene pool, thereby reducing genetic variation, or it may cause initially rare alleles, even neutral or deleterious ones, to become much more frequent or even fixed.
- genetic epidemiology
- The study of the role played by genetic factors in determining health and disease, in particular through the interaction of genetic factors with environmental factors, and typically as observed in genetically related individuals, often families or lineages but also populations and subpopulations.
- genetic erosion
- genetic genealogy
- The use of genealogical DNA testing in combination with traditional genealogical methods to infer the level and type of genetic relationships between individuals, to find ancestors, and to construct family trees, genograms, or other genealogical charts.
- genetic hitchhiking
- A type of linked selection by which the positive selection of an allele undergoing a selective sweep causes alleles for different genes at nearby loci to change frequency as well, allowing them to "hitchhike" to fixation along with the positively selected allele. If selection at the first locus is strong enough, neutral or even slightly deleterious alleles within the same linkage group may undergo the same positive selection because the physical distance between the nearby loci is small enough that a recombination event is unlikely to occur between them. Genetic hitchhiking is often considered the opposite of background selection.
- genetic load
- Any reduction in the mean fitness of a population owing to the existence of one or more genotypes with lower fitness than that of the most fit genotype.[1]
- genetic testing
- A broad class of procedures used to identify features of an individual's particular chromosomes, genes, or proteins in order to determine parentage or ancestry, diagnose vulnerabilities to heritable diseases, or detect mutant alleles associated with increased risks of developing genetic disorders. Genetic testing is widely used in human medicine, agriculture, and biological research.
- genetic variation
- The genetic differences both within and between populations, species, or other groups of organisms. It is often visualized as the variety of different alleles in the gene pools of different populations.
- genetic variability
- The formation or the presence of individuals differing in genotype within a population or other group of organisms, as opposed to individuals with environmentally induced differences, which cause only temporary, non-heritable changes in phenotype. Barring other limitations, a population with high genetic variability has a greater potential for successful adaptation to changing environmental conditions than a population with low genetic variability. Genetic variability is similar to, though distinct from, genetic diversity.
- genetic variation
- The genetic differences both within and between populations, species, or other groups of organisms. It is often visualized as the variety of different alleles in the gene pools of different populations.
- genetics
- The field of biology that studies genes, genetic variation, and heredity in living organisms.
- genic balance
- A mechanism of sex determination that depends upon the ratio of the number of X chromosomes (X) to the number of sets of autosomes (A). Males develop when the X/A ratio is 0.5 or less, females when it is 1.0 or more, and an intersex develops when it is between 0.5 and 1.0.[8]
- genic selection
- A type of natural selection that occurs at the level of individual genes or alleles, in which the frequency of an allele within a breeding population is determined by its fitness averaged over the variety of genotypes in which it occurs; the differential propagation of different alleles within a population as a consequence of properties borne by the alleles themselves, rather than by the genotypes in which they are found.[1]
- genic speciation
- genome
- The entire complement of genetic material contained within the chromosomes of an organism, organelle, or virus. The term is also used to refer to the collective set of genetic loci shared by every member of a population or species, regardless of the different alleles that may be present at these loci in different individuals.
- genomics
- An interdisciplinary field that studies the structure, function, evolution, mapping, and editing of entire genomes, as opposed to individual genes.
- genotype
- The entire complement of alleles present in a particular individual's genome, which gives rise to the individual's phenotype.
- genotype frequency
- The frequency or proportion of a population having a given genotype. Compare allele frequency.
- genotypic cluster species
- geographic speciation
- germ cell
- Any cell that gives rise to the gametes of an organism that reproduces sexually. Germ cells are the vessels for the genetic material which will ultimately be passed on to the organism's descendants and are usually distinguished from somatic cells, which are entirely separate from the germ line.
- germ line
- 1. In multicellular organisms, the population of cells which are capable of passing on their genetic material to the organism's progeny and are therefore (at least theoretically) distinct from somatic cells, which cannot pass on their genetic material except to their own immediate mitotic daughter cells. Cells of the germ line are called germ cells.
- 2. The lineage of germ cells, spanning many generations, that contains the genetic material which has been passed on to an individual from its ancestors.
- grade
- gradualism
- Continuous evolutionary change within a species lineage.[7] See also phyletic gradualism.
- green-beard effect
H
- habitat isolation
- Haldane's rule
- A rule formulated by J.B.S. Haldane which states that if one sex of the hybrid offspring resulting from a cross between two incipient species is inviable or sterile, that sex is more likely to be the heterogametic sex (i.e. the one with two different sex chromosomes).[18]
- haplodiploidy
- A sex-determination system in which the sex of an individual organism is determined by the number of sets of chromosomes it possesses: offspring which develop from fertilized eggs are diploid and female, while offspring which develop from unfertilized eggs are haploid and male, with half as many chromosomes as the females. Haplodiploidy is common to all members of the insect order Hymenoptera and several other insect taxa.
- haplogroup
- haploid
- (of a cell or organism) Having one copy of each chromosome, with each copy not being part of a pair. Contrast diploid and polyploid.
- haploinsufficiency
- haplotype
- A set of alleles in an individual organism that were inherited together from a single parent.
- Hardy–Weinberg principle
- A principle of population genetics which states that allele and genotype frequencies of a population will remain constant from generation to generation in the absence of other evolutionary influences. In the simplest case of a randomly mating population of diploid organisms possessing a single locus with two alleles, A and a, with frequencies f(A) = p and f(a) = q, respectively, the expected genotype frequencies are f(AA) = p2 for AA homozygotes, f(aa) = q2 for aa homozygotes, and f(Aa) = 2pq for heterozygotes. In the absence of evolutionary forces such as natural selection, mutation, assortative mating, gene flow, and genetic drift, p and q will remain constant between generations, such that the population is said to be in Hardy–Weinberg equilibrium with respect to the locus in question.
- hemizygous
- In a diploid organism, having just one allele at a given genetic locus (where there would ordinarily be two). Hemizygosity may be observed when only one copy of a chromosome is present in a normally diploid cell or organism, or when a segment of a chromosome containing one copy of an allele is deleted, or when a gene is located on a sex chromosome in the heterogametic sex (in which the sex chromosomes do not exist in matching pairs); for example, in human males with normal chromosomes, almost all X-linked genes are said to be hemizygous because there is only one X chromosome and few of the same genes exist on the Y chromosome.
- heredity
- The storage, transfer, and expression of genetic information in biological organisms,[13] as manifested by the passing on of phenotypic traits from parents to their offspring, either through sexual or asexual reproduction. Offspring cells or organisms are said to inherit the genetic information of their parents.
- heritability
- 1. The ability to be inherited.
- 2. A statistic used in quantitative genetics that estimates the proportion of variation within a given phenotypic trait that is due to genetic variation between individuals in a particular population. Heritability is estimated by comparing the individual phenotypes of closely related individuals in the population.
- heteropatric speciation
- heterosis
- The improved or increased function or quality of any biological trait in a hybrid offspring, with respect to the same trait in its genetically distinct parents. If any one or more of the parents' traits are noticeably enhanced in the offspring as a result of the mixing of the parents' genetic contributions, the offspring is said to be heterotic.
- heterozygous
- In a diploid organism, having two different alleles at a given genetic locus. In genetics shorthand, heterozygous genotypes are represented by a pair of non-matching letters or symbols, often an uppercase letter (indicating a dominant allele) and a lowercase letter (indicating a recessive allele), such as "Aa" or "Bb". Contrast homozygous.
- homoallele
- A mutant allele having a different mutation at the same site as another allele. Intragenic recombination between homoalleles cannot produce a functional cistron.[8]
- homologous chromosomes
- A set of two matching chromosomes, one maternal and one paternal, which pair up with each other inside the nucleus during meiosis. They have the same genes at the same loci, but may have different alleles.
- homology
- A similarity between a pair of structures, traits, or DNA sequences in different taxa that is due to shared ancestry.
- homoplasy
- homoploid recombinational speciation
- homozygous
- In a diploid organism, having two identical alleles at a given genetic locus. In genetics shorthand, homozygous genotypes are represented by a pair of matching letters or symbols, such as "AA" or "aa". Contrast heterozygous.
- horizontal gene transfer (HGT)
- Any process by which genetic material is transferred between unicellular and/or multicellular organisms other than by vertical transmission from parent to offspring, e.g. bacterial conjugation.
- host race
- host-specific parasite
- host-specific species
- hybrid
- The offspring that results from combining the qualities of two organisms of different genera, species, breeds, or varieties through sexual reproduction. Hybrids may occur naturally or artificially, as during selective breeding of domesticated animals and plants. Reproductive barriers typically prevent hybridization between distantly related organisms, or at least ensure that hybrid offspring are sterile, but fertile hybrids may result in speciation.
- hybrid breakdown
- hybrid incompatibility
- hybrid inviability
- hybrid speciation
- hybrid sterility
- hybrid swarm
- hybrid zone
- A geographic area in which the ranges of two interbreeding species or populations overlap, allowing them to cross-fertilize and generate hybrid offspring. The formation of a hybrid zone is one of the four outcomes of secondary contact between divergent genetic lineages.
- hybridization
- The process by which a hybrid organism is produced from two parents of different genera, species, breeds, or varieties.
- hypermorphosis
- The exaggeration of one or more phenotypic features of a descendant organism compared to those of its ancestors due to an increase in the duration of ontogenetic development over evolutionary history.[1]
I
- identical ancestors point
- identical by descent (IBD)
- (of a gene or allele) Traceable back through an arbitrary number of generations without mutation to a common ancestor of the group of descendant organisms that carries the gene or allele.[1] A gene or allele present in a group of descendant organisms is said to be identical by descent to a gene or allele in a common ancestor of the group if both sequences are identical, indicating that the sequence has been passed down unmodified from the common ancestor to its descendants.
- inbred line
- Any lineage of a particular species in which individuals are nearly or completely genetically identical to each other due to a long history of repeated inbreeding, either by natural or artificial means. Lineages are typically considered inbred after at least 20 generations of inbreeding (e.g. by self-fertilization or sib mating), at which point nearly all loci across the genome are homozygous and all individuals can therefore effectively be treated as clones (despite the fact that individuals are still produced by sexual reproduction).
- inbreeding
- Sexual reproduction between breeds or individuals that are closely related genetically. Inbreeding results in homozygosity, which can increase both the probability of offspring being affected by deleterious recessive traits and the probability of fixing beneficial traits within the breeding population. The reproductive event and the resulting progeny may both be referred to as an incross, and the progeny is said to be inbred. Contrast outbreeding.
- inbreeding depression
- inclusive fitness
- The number of offspring equivalents that an individual organism rears, rescues, or otherwise supports through its behavior, regardless of whether or not the individual is actually a biological parent of the offspring equivalents. Inclusive fitness is one of two metrics of evolutionary success as defined by W.D. Hamilton in 1964, the other being personal fitness.
- incidence
- The frequency of new occurrence of a genetic disorder (or more broadly any genetic condition or trait, deleterious or otherwise) among the members of a particular population and within a particular period of time.[13]
- incomplete dominance
- incomplete speciation
- incipient species
- Any population that is in an early stage of speciation.
- inheritance
- See heredity.
- interbreeding
- intercross
- A cross in which both the male and female parents are heterozygous at a particular locus.[8]
- intrinsic postzygotic isolation
- introgression
- The movement of a gene from the gene pool of one population or species into that of another population by the repeated backcrossing of hybrids of the two populations with one of the parent populations. Introgression is a ubiquitous and important source of genetic variation in natural populations, but may also be practiced intentionally in the cultivation of domesticated plants and animals.
- inviability
- isolating mechanism
- isolation
- isolation by distance
- isomeric genes
- Two or more genes that are equivalent and redundant in the sense that, despite coding for distinct gene products, they each result in the same phenotype when set within the same genetic background. If several isomeric genes are present in a single genotype they may be either cumulative or non-cumulative in their contributions to the phenotype.[13]
- iterative evolution
- The repeated evolution of similar phenotypic characteristics or traits in different organisms at different times during the evolutionary history of a clade,[1] a phenomenon which can result in the seeming de-extinction of an organism previously considered extinct.
- iteroparity
- A reproductive strategy characterized by multiple reproductive cycles during an individual organism's lifetime. Organisms that use such a strategy are said to be iteroparous. Iteroparity is usually contrasted with semelparity.
J
- Jordan's Law
K
- K-strategist
- Ka/Ks ratio
- Kaneshiro model
- A model of peripatric speciation developed by Kenneth Y. Kanneshiro where a sexual species experiences a population bottleneck—that is, when the genetic variation is reduced due to small population size—mating discrimination among females may be altered by the decrease in courtship behaviors or displays of males. This allows sexual selection to give rise to novel sexual traits in the new population.[19]
- kin
- kin recognition
- kin selection
- A form of genic selection by which alleles differ in their rates of propagation by influencing the survival or reproductive success of individuals who carry the same alleles by common descent (their kin).[1]
- koinophilia
- An evolutionary hypothesis which proposes that during sexual selection, organisms preferentially seek mates with a minimum of unusual or mutant traits, e.g. in terms of functionality, appearance, or behavior. The hypothesis attempts to explain the clustering of sexual organisms into distinct species and other issues described by Darwin's dilemma.
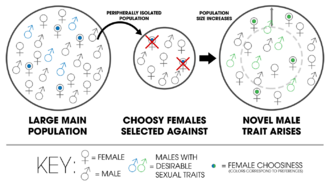
In the Kaneshiro model of peripatric speciation, a sample of a larger population results in an isolated population with less males containing attractive traits. Over time, choosy females are selected against as the population increases. Sexual selection drives new traits to arise (green), thereby reproductively isolating the new population from the old one (blue).
L
- last universal common ancestor (LUCA)
- The most recent population of organisms from which all extant organisms on Earth share a common descent; i.e. the most recent common ancestor of all organisms now living. LUCA is not thought to have been the earliest life on Earth, but rather the only organism of its time to still have living descendants. Its existence is not known from any specific fossil record but is inferred from phylogenetic comparisons of modern organisms, all of which are its descendants.
- lateral gene transfer (LGT)
- See horizontal gene transfer.
- Law of Dominance and Uniformity
- One of three fundamental principles of Mendelian inheritance, which states that different alleles of the same gene may be dominant or recessive relative to others, and that an organism with at least one dominant allele will uniformly display the phenotype associated with the dominant allele.
- Law of Independent Assortment
- One of three fundamental principles of Mendelian inheritance, which states that genes responsible for different phenotypic traits are segregated independently during meiosis. Linked genes are a notable exception to this rule.
- Law of Segregation
- One of three fundamental principles of Mendelian inheritance, which states that during meiosis, the alleles of each gene segregate from each other such that each resulting gamete carries only one allele of each gene.
- lineage
- A linear evolutionary sequence connecting an ancestral cell, organism, or species to a particular descendant cell, organism, or species, including all intermediate organisms and spanning any number of generations; the direct progression of reproductive events (i.e. the line of descent) between two individuals, including vertically related individuals, e.g. parent(s) and offspring, but usually excluding horizontally related individuals who did not themselves directly contribute genetic material to any of the included individuals, e.g. siblings.
- lineage-splitting
- When gene flow between two populations is completely eliminated.[7]
- linkage
- The tendency of DNA sequences which are physically near to each other on the same chromosome to be inherited together during meiosis. Because the physical distance between them is relatively small, the chance that any two nearby parts of a DNA sequence (often loci or genetic markers) will be separated on to different chromatids during chromosomal crossover is statistically very low; such loci are then said to be more linked than loci that are farther apart. Loci that exist on entirely different chromosomes are said to be perfectly unlinked. The standard unit for measuring genetic linkage is the centimorgan (cM).
- linkage disequilibrium
- locus
- A specific, fixed position on a chromosome where a particular gene or genetic marker resides.
- lyonization
- See X-inactivation.
M
- macroevolution
- Evolutionary change as it occurs at a relatively large scale, at or above the level of species, as opposed to microevolution, which occurs at a smaller scale. Macroevolution is often thought of as the compounded effects of microevolution.
- map unit (m.u.)
- See centimorgan.
- maternal effect
- Any nongenetic effect of the mother on the phenotype of her offspring, owing to factors such as cytoplasmic inheritance, transmission of congenital disease, and the sharing of nutritional conditions.[1]
- mating system
- mating system isolation
- matroclinous
- (of an offspring) Resembling the female parent, genotypically or phenotypically, more closely than the male parent; derived from the mother. Contrast patroclinous.
- maximum parsimony
- See parsimony.
- mechanical isolation
- mechanical pollinator isolation
- meiosis
- A specialized type of cell division that occurs exclusively in sexually reproducing eukaryotes, during which DNA replication is followed by two consecutive rounds of division to ultimately produce four genetically unique haploid daughter cells, each with half the number of chromosomes as the original diploid parent cell. Meiosis only occurs in cells of the sex organs, and serves the purpose of generating haploid gametes such as sperm, eggs, or spores, which are later fused during fertilization. The two meiotic divisions, known as Meiosis I and Meiosis II, may also include various genetic recombination events between homologous chromosomes.
- Mendelian inheritance
- A theory of biological inheritance based on a set of principles originally proposed by Gregor Mendel in 1865 and 1866. Mendel derived three generalized laws about the genetic basis of inheritance which, together with several theories developed by later scientists, are considered the foundation of classical genetics. Contrast non-Mendelian inheritance.
- meristic trait
- A discretely varying, countable trait, e.g. number of digits.[1]
- metagenomics
- The study of genetic material recovered directly from environmental samples, as opposed to organisms cultivated in laboratory cultures.
- microallopatric
- Allopatric speciation occurring on a small geographic scale.[20]
- microevolution
- Evolutionary change as it occurs at a relatively small scale, typically within a particular species or population, as opposed to macroevolution, which occurs at a larger scale. Because of the convenience of observing and modeling small-scale changes in allele frequencies within discrete populations, the principles of population genetics are often conceptualized at microevolutionary scales.
- microspecies
- midparent value
- The mean of the two parental values for a quantitative trait in an individual offspring or in a specific cross.[8]
- migration
- mimicry
- The process by which an organism evolves to resemble another object, often an organism of another species. Mimicry can also occur between individuals of the same species. A type of adaptive signaling, mimicry evolves when a signal-receiver, known as the dupe, perceives the similarity between the mimic and the object or organism it is mimicking, known as the model, and as a result changes its behavior in a way that provides a selective advantage to the mimic; the model may also benefit from the shared resemblance, in which case there is a mutualism, or the mimicry may be to the model's detriment, making it parasitic or competitive. The evolved resemblance may be visual, acoustic, chemical, tactile, or electrical, or any combination of sensory modalities. There are many varieties of mimicry, such as Batesian, Müllerian, and Vavilovian.
- minor allele frequency (MAF)
- missense mutation
- A type of point mutation which results in a codon that codes for a different amino acid than in the unmutated sequence. Compare nonsense mutation.
- mitochondrial Eve
- mitosis
- In eukaryotic cells, the part of the cell cycle during which the division of the nucleus takes place and replicated chromosomes are separated into two distinct nuclei. Mitosis is generally preceded by the S stage of interphase, when the cell's DNA is replicated, and either occurs simultaneously with or is followed by cytokinesis, when the cytoplasm and cell membrane are divided into two new daughter cells.
- modern synthesis
- modes of speciation
- A classification scheme of speciation processes based on the level of gene flow between two populations.[21] The traditional terms for the three modes—allopatric, parapatric, and sympatric—are based on the spatial distributions of a species population.[22][21]
- molecular genetics
- A branch of genetics that employs methods and techniques of [[Glossary of genetics (M–Z)molecular biology#{{{1}}}|{{{1}}}]] to study the structure and function of genes and gene products at the molecular level. Contrast classical genetics.
- monophyly
- morphological species concept
- mosaic evolution
- The evolutionary change of certain adaptive structures, traits, or other components of the phenotype at different times or different rates than others, either within a single species or between different species.[23]
- mosaic hybrid zone
- A zone in which two speciating lineages occur together in a patchy distribution–either by chance, random colonization, or low hybrid fitness.[21]
- mosaic sympatry
- A case of sympatry in which two populations overlapping in geographic distribution exhibit habitat specializations.[21]
- mosaicism
- The presence of two or more populations of cells with different genotypes in an individual organism which has developed from a single fertilized egg. A mosaic organism can result from many kinds of genetic phenomena, including nondisjunction of chromosomes, endoreduplication, or mutations in individual stem cell lineages during the early development of the embryo. Mosaicism is similar to but distinct from chimerism.
- most recent common ancestor (MRCA)
- Muller's ratchet
- multifurcation
- See polytomy.
- mutant
- An organism, gene product, or phenotypic trait resulting from a mutation, of a type that would not be observed naturally in wild-type specimens.
- mutation
- Any permanent change in the nucleotide sequence of a strand of DNA or RNA, or in the amino acid sequence of a peptide. Mutations play a role in both normal and abnormal biological processes; their natural occurrence is integral to the process of evolution. They can result from errors in replication, chemical damage, exposure to high-energy radiation, or manipulations by mobile genetic elements. Repair mechanisms have evolved in many organisms to correct them. By understanding the effect that a mutation has on phenotype, it is possible to establish the function of the gene or sequence in which it occurs.
- mutation distance
- The smallest number of mutations required to derive one particular gene, sequence, or phenotype from another;[8] the minimum number of nucleobase insertions, deletions, or substitutions necessary to change one sequence into another.
- mutation event
- The actual origin of a particular mutation in time and space; the instance of its original introduction into a genome, as opposed to that of its phenotypic manifestation, which may only occur generations after the fact.
- mutation rate
- The frequency of new mutations at a particular locus or in a particular gene, sequence, genome, or organism over a specified period of time, e.g. during a single generation. Mutation rates may be calculated for a specific class of mutation or for all types collectively; they vary widely by organism and with an organism's environment.
- mutational hot spot
- mutational load
- See genetic load.
- mutational meltdown
- mutationism
- mutator gene
- Any mutant gene or sequence that increases the spontaneous mutation rate of one or more other genes or sequences. Mutators are often transposable elements, or may be mutant housekeeping genes such as those that encode helicases or proteins involved in proofreading.[8]
N
- natural selection
- neontology
- The study of extant taxa, i.e. those with members that are still living in the present day, as opposed to paleontology.
- network evolution
- See reticulate evolution.
- neutral mutation
- 1. Any mutation of a nucleic acid sequence that is neither beneficial nor detrimental to the ability of an organism to survive and reproduce.
- 2. Any mutation for which natural selection does not affect the spread of the mutation within a population.
- nexus hypothesis
- The hypothesis that each phenotypic trait is likely to be influenced by more than one gene, and conversely that most genes affect more than one phenotype.[23]
- niche
- 1. The ecological role of a particular species or other taxon in a larger community, generally conceptualized as the multidimensional space, of which the coordinates are the various parameters representing the conditions which are necessary for the existence of the species in every aspect of its present form, to which a species is restricted by the presence of competitor species.[23]
- 2. A particular environment or environmental condition to which a species is matched; the variety of activities, behaviors, and ecological functions carried out by an organism or population in response to its environmental context, e.g. the distribution of resources and competitors, and the ways in which it in turn alters that same context. The term is sometimes used loosely as an equivalent of microhabitat, in the sense of the physical space occupied by a species.[23] See also fundamental niche and realized niche.
- niche adaptation
- niche preference
- noncompetitive gametic isolation
- nondisjunction
- The failure of homologous chromosomes or sister chromatids to segregate properly during cell division. Nondisjunction results in daughter cells that are aneuploid, containing abnormal numbers of one or more specific chromosomes. It may be caused by any of a variety of factors.
- nongenetic barrier
- nonhomologous recombination
- nonreciprocal recombination
- See unequal crossing over.
- nonsense mutation
- A type of point mutation which results in a premature stop codon in the transcribed mRNA sequence, thereby causing the premature termination of translation, which results in a truncated, incomplete, and often non-functional protein.
- nonsynonymous mutation
- A type of mutation in which the substitution of one nucleotide base for another results, after transcription and translation, in an amino acid sequence that is different from that produced by the original unmutated gene. Because nonsynonymous mutations always result in a biological change in the organism, they are often subject to strong selection pressure. Contrast synonymous mutation.
- non-geographic speciation
- non-Mendelian inheritance
- Any pattern of inheritance in which traits do not segregate in accordance with Mendel's laws, which describe the readily observable inheritance of discretely variable phenotypic traits influenced by single genes located on nuclear chromosomes. Though they correctly explain many basic observations of inheritance, Mendel's laws are useful only in the simplest and most general cases; there exist numerous genetic processes and phenomena, both normal and abnormal, which violate them, such as incomplete dominance, codominance, genetic linkage, epistatic interactions and polygenic traits, non-random segregation of chromosomes, extranuclear inheritance, gene conversion, and many epigenetic phenomena.
- non-random segregation of chromosomes
- norm of reaction
- See reaction norm.
- null allele
- Any allele made non-functional by way of a genetic mutation. The mutation may result in the complete failure to produce a gene product or a gene product that does not function properly; in either case, the allele may be considered non-functional.
- nullisomy
- The condition of a cell or organism lacking all of the copies of a particular chromosome that are normal for its ploidy level; e.g. in a diploid organism, lacking both members of the normal pair. Nullisomy is frequently lethal early in development.
- nullizygous
O
- offspring
- ontogeny
- The origination and biological development of an organism within its own lifetime, as opposed to phylogeny, which refers to the evolutionary history of the organism's ancestors. In sexually reproducing organisms, ontogeny is the study of the development of an organism from the time of fertilization to the organism's reproductively mature form; the term may also be used to refer to the study of an organism's entire lifespan.
- operational taxonomic unit (OTU)
- orphan gene
- A gene for which there are no known functional homologs outside of a given species or lineage, and whose evolutionary history is therefore obscure.
- orphon
- Any coding or non-coding DNA sequence which is derived from a tandem multigene family or cluster but is physically isolated from the other genes in the family because it is dispersed to a distant locus in the genome. Orphons are usually non-functional pseudogenes with highly variable copy numbers.[24]
- orthogenesis
- ortholog
- One of a set of genes (or more generally any DNA sequences showing homology) which are present in different genomes but are descended from the same ancestral sequence, i.e. they are directly related to one another by vertical descent from a single gene or sequence in the most recent common ancestor of those genomes. Such genes or sequences are said to be orthologous. Orthologs can be inferred to be related to each other based on the similarity of their sequences; though they may have evolved independently within separate genomes by mutation and natural selection, their products may still retain similar structures, functions, or levels of expression across species and populations. The identification of orthologs has proven important in inferring phylogenetic relationships between organisms. Contrast paralog.
- outbreeding
- Sexual reproduction between different breeds or individuals, which has the potential to increase genetic diversity by introducing unrelated genetic material into a breeding population. The reproductive event and the resulting progeny may both be referred to as an outcross, and the progeny is said to be outbred. Contrast inbreeding.
- outgroup
P
- paleontology
- paleopolyploidy
- para-allopatric speciation
- A mode of speciation in which divergence begins in parapatry but is completed in allopatry.[3]
- paracentric inversion
- A chromosomal inversion in which the inverted segment does not include the chromosome's centromere. Contrast pericentric inversion.
- parallel evolution
- The independent evolution of similar or identical derived traits or characters in related lineages, thought usually to be based on similar modifications of common developmental pathways.[1] Contrast convergent evolution.
- parallel speciation
- paralog
- One of a set of genes (or, more generally, any DNA sequences showing homology) which are directly related to each other via one or more genetic duplication events; such genes or sequences are said to be paralogous. Paralogs result from the duplication of a single sequence within a single genome and then the subsequent divergence of the duplicated sequences by mutation and natural selection (either within the original genome, or, during speciation, in different genomes). Contrast ortholog.
- parapatric speciation
- paraphyly
- parasitic DNA
- See selfish genetic element.
- parsimony
- The principle of accounting for empirical observations by whichever hypothesis requires the fewest or the simplest assumptions for which there is limited or no evidence. In biological systematics, maximum parsimony is an optimality criterion which invokes a minimum of evolutionary changes to infer phylogenetic relationships; i.e. the phylogenetic tree that minimizes the total number of character-state changes is to be preferred.[1]
- parthenogenesis
- A type of asexual reproduction in which the growth and development of embryos occurs without fertilization. In animals which reproduce by parthenogenesis, an unfertilized gamete of the female parent is capable of developing into an adult without any contribution from a male parent, resulting in offspring possessing only the mother's genetic material (the exact proportion of which depends on the parthenogenetic mechanism, of which there are numerous varieties). Some species reproduce exclusively by parthenogenesis, while others can switch between sexual reproduction and parthenogenesis under certain environmental conditions.
- partial dominance
- See incomplete dominance.
- particulate inheritance
- One of the defining ideas of Mendelian inheritance, which holds that phenotypic traits are or can be inherited via the passing of "discrete particles" from generation to generation. These particles may not have detectable effects in every generation but nevertheless retain their ability to be expressed in subsequent generations.
- patroclinous
- (of an offspring) Resembling the male parent, genotypically or phenotypically, more closely than the female parent;[8] derived from the father. Contrast matroclinous.
- peak shift model
- pedigree chart
- penetrance
- The proportion of individuals with a given genotype who express the associated phenotype, usually given as a percentage. Because of the many complex interactions that govern gene expression, the same allele may produce an observable phenotype in one individual but not in another. If less than 100% of the individuals in a population carrying the genotype of interest also express the associated phenotype, both the genotype and phenotype may be said to show incomplete penetrance. Penetrance quantifies the probability that an allele will result in the expression of its associated phenotype in any form, i.e. to any extent that makes an individual carrier different from individuals without the allele. Compare expressivity.
- pericentric
- (of a gene or region of a chromosome) Positioned near the centromere of the chromosome.
- pericentric inversion
- A chromosomal inversion in which the inverted segment includes the chromosome's centromere. Contrast paracentric inversion.
- peripatric speciation
- A variation of allopatric speciation where a new species forms from a small, peripheral isolated population.[25] It is sometimes referred to as centripetal speciation in contrast to centrifugal speciation.
- phenetic
- Pertaining to phenotypic similarity, e.g. a phenetic classification.[1]
- phenome
- The complete set of phenotypes that are or can be expressed by a genome, cell, tissue, organism, or species; the sum of all of its manifest chemical, morphological, and behavioral characteristics or traits.
- phenomic lag
- A delay in the phenotypic expression of a genetic mutation owing to the time required for the manifestation of changes in the affected biochemical pathways.[13]
- phenotype
- The composite of the observable morphological, physiological, and behavioral traits of an organism that result from the expression of the organism's genotype as well as the influence of environmental factors and the interactions between the two.
- phyletic gradualism
- A model of evolution which theorizes that most speciation occurs slowly, uniformly, and gradually, and that there is seldom a clear line of demarcation between ancestral species and descendant species unless there is a sudden split which reproductively isolates members of the same population. The theory is often contrasted with punctuated equilibrium.
- phylogenetic bracketing
- A method used to infer the likelihood of specific traits being present in organisms whose phenotypes are incomplete or unknown based on their positions in a phylogenetic tree relative to ancestors, descendants, or contemporaneous organisms with more completely understood phenotypes. A major application of this method is in paleontology, where extinct organisms known only from fossils are compared to their closest known relatives in order to infer the presence or absence of certain traits for which fossils provide limited or no evidence, such as soft tissues, integumentary structures, and physiological and behavioral traits, though the method is extremely sensitive to confounds from convergent evolution.
- phylogenetic species concept
- phylogenetic tree
- A graphical representation of a phylogeny, consisting of a branching, tree-like diagram showing the evolutionary relationships between biological species or other taxa as inferred from similarities and differences in their morphological or genetic characteristics, and how they have all descended from a common ancestor.
- phylogenetics
- The study of the evolutionary history of and relationships between individuals or groups of organisms, such as species or populations, through methods that evaluate similarities and differences between observed heritable traits, including morphological features and DNA sequences. The graphical presentation of data from such analyses is known as a phylogeny or phylogenetic tree.
- phylogeny
- phylogeography
- pleiotropy
- The phenomenon by which one gene influences two or more seemingly unrelated phenotypic traits, by any of several distinct but potentially overlapping mechanisms.
- plesiomorphy
- An ancestral character state; i.e. the state or condition of a particular trait or feature (e.g. a specific phenotype) that is present in the common ancestor of a given clade. Plesiomorphies may or may not be shared by some or all descendants within the clade. The term is relative; a trait considered a plesiomorphy in one clade may not be considered a plesiomorphy in a different clade. Contrast apomorphy.
- ploidy
- The number of complete sets of chromosomes in a cell, and hence the number of possible alleles present within the cell at any given autosomal locus.
- pollinator isolation
- polygene
- polygenic trait
- Any phenotypic trait which is under the direct control of more than one gene. Polygenic traits are often quantitative traits.
- polymorphism
- The regular and simultaneous occurrence in the same population of two or more alleles (or genotypes) at the same locus at frequencies that cannot be accounted for by recurrent mutation alone (generally at least 1%), implying that the multiple alleles are being stably inherited by members of the population.[13]
- polypheny
- See pleiotropy.
- polyphyly
- The grouping of organisms which do not share an immediate common ancestor; such groups are said to be polyphyletic. The term is often applied to groups of organisms that share characteristics which appear to be similar but are not actually closely related, frequently as a result of convergent evolution. The avoidance of polyphyletic groupings is often a stimulus for major revisions of biological classification schemes. Contrast monophyly and paraphyly.
- polyploid
- (of a cell or organism) Having more than two homologous copies of each chromosome. Polyploidy may occur as a normal condition of chromosomes in certain cells or even entire organisms, or it may result from abnormal cell division or a mutation causing the duplication of the entire chromosome set. Contrast haploid and diploid; see also ploidy.
- polytomy
- popular sire effect
- population
- A group of organisms of the same species which occupies a more or less well-defined geographic region and which exhibits reproductive continuity from generation to generation. It is generally presumed that ecological and reproductive interactions occur more frequently among the members of the group than between them and members of other populations of the same species.[1]
- population bottleneck
- A sharp, often sudden reduction in the size of a biological population, often due to a major environmental event such as a flood, fire, volcanic eruption, drought, famine, or disease. Because only a small population with a narrower range of genetic diversity remains afterward to pass on genes to future generations, such events tend to reduce the genetic variation in the population's gene pool, and often lead to new and distinct populations through founder effects. Diversity increases again only when gene flow from another population occurs, or very slowly over time as random mutations accumulate.
- population genetics
- A subfield of genetics and evolutionary biology that studies genetic differences within and between populations of organisms.
- position effect
- Any effect on the expression or functionality of a gene or sequence that is a consequence of its location or position within a chromosome or other DNA molecule. A sequence's precise location relative to other sequences and structures tends to strongly influence its activity and other properties, because different loci on the same molecule can have substantially different genetic backgrounds and physical/chemical environments, which may also change over time. For example, the transcription of a gene located very close to a nucleosome, centromere, or telomere is often repressed or entirely prevented because the proteins that make up these structures block access to the DNA by transcription factors, while the same gene is transcribed at a much higher rate when located in euchromatin. Proximity to promoters, enhancers, and other regulatory elements, as well as to regions of frequent transposition by mobile elements, can also directly affect expression; being located near the end of a chromosomal arm or to common crossover points may affect when replication occurs and the likelihood of recombination. Position effects are a major focus of research in the field of epigenetic inheritance.
- positive selection
- See directional selection.
- postmating barrier
- postmating prezygotic isolation
- postzygotic isolation
- preadaptation
- Possession of the necessary properties to permit a shift into a new niche or habitat. A structure is said to be preadapted if it can assume a new function before it itself becomes modified by selection.[1]
- premating barrier
- premating isolation
- prezygotic isolation
- proband
- A term used in medical genetics and genealogy to denote a particular subject being studied or reported on.
- progenesis
- The precocious or accelerated sexual maturation of an organism that is still at a morphologically juvenile stage.[23]
- progeny
- A genetic descendant or group of descendants; the offspring of a single reproductive event, either sexual or asexual.[23]
- progressive selection
- See directional selection.
- protosexual
- Of or pertaining to organisms that achieve genetic recombination by conjugation, transduction, or lysogenization.[23] Compare eusexual and parasexual.
- protospecies
- An ancestral species.[23]
- protype
- In taxonomy, a complete specimen that replaces a fragmentary holotype.[23]
- pseudoallele
- Any of two or more different genes or sequences which have the same or similar contributions to phenotype, and thus appear to be genuine alleles, but are not actually structurally allelic (i.e. they do not occupy homologous loci on homologous chromosomes).[23]
- pseudogene
- A non-functional sequence of DNA that resembles a functional gene. Pseudogenes are typically superfluous copies of functional genes which have been duplicated by natural processes, except that they lack regulatory sequences necessary for proper transcription or translation or contain other defects such as frameshift mutations, premature stop codons, or missing introns.
- pseudopolyploidy
- 1. The condition in which the number of chromosomes in a chromosome set is doubled (or tripled, etc.) but without a corresponding increase in the actual amount of genetic material (i.e. the ploidy level). This occurs when the chromosomes of a normal chromosome complement (e.g. diploid) become fragmented into smaller pieces, increasing the total number of individual chromosomes but not creating additional homologous copies of those chromosomes (such that the cell remains diploid).[23]
- 2. Any numerical relationship between chromosome sets in groups of related organisms which suggests that some of those organisms are polyploids of others when in fact they are not.[23]
- punctuated equilibrium
- Punnett square
- A tabular diagram used to predict the possible genotypes that can be inherited by the offspring of a particular cross or breeding experiment by summarizing all of the various combinations of maternal alleles with paternal alleles. The resulting table can then be used to determine the probabilities that the offspring will have a particular genotype. The usefulness of Punnett squares is limited to discrete phenotypes inherited according to simple Mendelian patterns.
- purebred
- putative gene
- A specific nucleotide sequence suspected to be a functional gene based on the identification of its open reading frame. The gene is said to be "putative" in the sense that no function has yet been described for its products.

Parapatric speciation can occur when the members of a population subject to a selective gradient of phenotypic or genotypic frequencies (a cline) experience different selective conditions at each end of the gradient (divergent selection). Reproductive isolation occurs upon the formation of a hybrid zone. In most cases, the hybrid zone is eliminated due to a selective disadvantage, which effectively completes the speciation process.
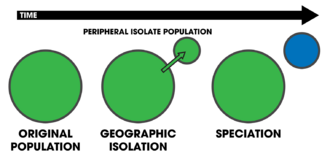
In peripatric speciation, a small population becomes isolated on the periphery of the central population evolving reproductive isolation (blue) due to reduced gene flow.
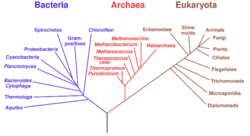
A phylogenetic tree depicting the evolutionary relationships between the three domains of life (Bacteria, Archaea, and Eukaryota) and the major clades within them. The root of the tree symbolizes that all extant life on Earth descended from a single common ancestor.
Q
- quantitative genetics
- A branch of population genetics which studies phenotypes that vary continuously (such as height or mass) as opposed to those that fall into discretely identifiable categories (such as eye color or the presence or absence of a particular trait). Quantitative genetics employs statistical methods and concepts to link continuously distributed phenotypic values to specific genotypes and gene products.
- quantitative trait
- quantitative trait locus (QTL)
- quantum evolution
- A rapid evolutionary shift in a lineage to a phenotypic state that is distinctly unlike the ancestral condition.[1]
- quantum speciation
- A chromosomal model of speciation that occurs rapidly when a cross-fertilizing plant species buds off from a larger population on the periphery, experiencing interbreeding and strong genetic drift that results in a new species.[26][27][28] The model is similar to that of Ernst Mayr's peripatric speciation.[29]
R
- r/K selection
- The natural selection of combinations of traits in organisms or species which appear to involve a trade-off between quantity and quality of offspring, whereby an organism or species may evolve to make use of either of two different reproductive strategies: r-strategists tend to produce many, low-quality offspring, yielding large numbers of progeny during their lifespan but investing little or no energy in nurturing or protecting them, whereas K-strategists tend to produce few, high-quality offspring, yielding small numbers of progeny but with a corresponding increase in parental investment. Which strategy evolves depends on which one results in greater reproductive success, which itself often depends on the stability of the organism's environment. In an unstable environment, where the probability that any individual offspring will survive to maturity is low, investment in parental care may not be sensible, and the parent may be more likely to pass on its genetic material if it dedicates its metabolic energy to simply producing as many offspring as possible rather than to parenting. Conversely, in more stable environments where survival to maturity is relatively common, the parent may find greater success if it dedicates more time and energy to parental care, improving each individual offspring's likelihood of reproducing successfully. The different strategies are often accompanied by characteristic anatomical or physiological traits, e.g. r-selected species often have small body size, rapid development, and short lifespans.
- reaction norm
- The pattern or set of phenotypic expressions of a given genotype across a variety of different environmental conditions.[1]
- recapitulation
- The ontogenetic passage of an organism's features through stages that resemble the adult features of the organism's phylogenetic ancestors.[1]
- recessiveness
- A relationship between the alleles of a gene in which one allele produces an effect on phenotype that is overpowered or "masked" by the contribution of another allele at the same locus; the first allele and its associated phenotypic trait are said to be recessive, and the second allele and its associated trait are said to be dominant. Often, recessive alleles code for inefficient or dysfunctional proteins. Like dominance, recessiveness is not an inherent property of any allele or phenotype, but simply describes its relationship to one or more other alleles or phenotypes. In genetics shorthand, recessive alleles are often represented by a lowercase letter (e.g. "a", in contrast to the dominant "A").
- reciprocal cross
- A crossbreeding experiment designed to test whether parental sex influences the inheritance of a particular trait. In crosses where the parents differ in genotype or phenotype or both, and hence only one of the parents (either the male or female parent) expresses the trait of interest, the reciprocal cross is the inverse, in which the parent of the other sex expresses the trait of interest instead. For example, if in the first cross a male expressing the trait is crossed with a female not expressing it, then in the reciprocal cross a female expressing the trait is crossed with a male not expressing it. By observing the progeny resulting from each cross, geneticists can make inferences about which sex chromosome, if either, influences the trait's expression.
- reciprocal hybrid
- A hybrid offspring resulting from a reciprocal cross between parents differing in genotype or phenotype or both.[8]
- reciprocal translocation
- A type of chromosomal translocation by which there is a reciprocal exchange of chromosome segments between two or more non-homologous chromosomes. When the exchange of material is evenly balanced, reciprocal translocations are usually harmless.
- recognition species concept
- recombinational speciation
- recurrent evolution
- The repeated evolution of a particular trait or character, for whatever reason, whether by natural selection or genetic drift.
- Red Queen hypothesis
- refugium
- A geographic location (or, more narrowly, a niche) in which one or more species has persisted while becoming extinct elsewhere.[1]
- reinforcement
- A process of speciation by which natural selection increases the reproductive isolation between two populations of a species as a result of selection acting against the production of hybrid individuals of low fitness.[3] See also Evidence of speciation by reinforcement.
- relict
- A species or population that is the last surviving representative of an otherwise extinct group, taxon, lineage, or clade, or which has been left behind in a locality after extinction throughout most of a formerly larger geographic distribution.[1]
- repression
- See downregulation.
- reproduction curve
- A graphical representation of the relationship between the number of individuals at a given stage of one generation and the number of individuals at the same stage in a previous generation.[23]
- reproductive character displacement
- reproductive effort
- The proportion of an individual's total metabolic resources that is devoted to reproduction.[23]
- reproductive isolating barriers
- The set of evolutionary mechanisms, behaviors, and physiological processes responsible for the reproductive isolation of two or more populations.
- reproductive isolation
- The condition in which interbreeding between two or more populations of organisms is prevented by intrinsic factors, such that the members of one population cannot mate with the members of another population and produce fertile offspring. The evolution of reproductive isolation between members of different populations is usually considered the first step in the process of speciation, because it effectively prevents gene flow between the populations and thereby allows each to evolve independently; hence the existence of reproductive barriers is often used as a criterion by which to define species in various species concepts. Isolation may occur when the populations are physically separated by environmental changes or migration such that members of the other population are simply inaccessible, or it may occur when anatomical or genetic differences make copulation between members of different populations impossible or at least ensure that any offspring that happen to develop are sterile, even though the populations are not physically separated from each other. Isolating mechanisms are typically classified as prezygotic (isolating barriers occurring before the formation of a zygote) and postzygotic (isolating barriers occurring after the formation of a zygote).
- reproductive success
- The successful production of offspring by an individual, often quantified as the number of offspring produced by the individual per reproductive event or during the individual's entire lifespan, or as the number of an individual's offspring that survive to reproductive maturity themselves or that are surviving at a given time.[23]
- reproductivity effect
- The decrease in the rate of reproduction of new individuals per colony member as colony size increases.[23]
- reticulate evolution
- The union of different lineages of a clade by hybridization.[1]
- reverse genetics
- An experimental approach in molecular genetics in which a researcher starts with a known gene and attempts to determine its function or its effect on phenotype by any of a variety of laboratory techniques, commonly by deliberately mutating the gene's nucleic acid sequence or by repressing or silencing its expression and then screening the mutated organisms for obvious changes in phenotype. When the gene of interest is the only one in the genome whose expression has been manipulated, any observed phenotypic changes are assumed to be influenced by it. This is the opposite of forward genetics, in which a known phenotype is linked to one or more unknown genes.
- reverse mutation
- Any mutation in a gene or DNA sequence which restores or rescues the original function or phenotype that was altered or destroyed by a previous mutation in the same sequence.[8] Contrast forward mutation; see also suppressor mutation.
- revertant
- A gene or allele in which a reverse mutation occurs, or an organism bearing such a gene or allele.[8]
- ring species
- Connected populations of the same species, each of which can interbreed with closely sited, closely related populations, but for which there exist at least two "end" populations in the series which are too distantly related to interbreed.
- Robertsonian translocation (ROB)
- A type of chromosomal translocation by which double-strand breaks at or near the centromeres of two acrocentric chromosomes cause a reciprocal exchange of segments that gives rise to one large metacentric chromosome (composed of the long arms) and one extremely small chromosome (composed of the short arms), the latter of which is often subsequently lost from the cell with little effect because it contains very few genes. The resulting karyotype shows one fewer than the expected total number of chromosomes, because two previously distinct chromosomes have essentially fused together. Carriers of Robertsonian translocations are generally not associated with any phenotypic abnormalities, but do have an increased risk of generating meiotically unbalanced gametes.
- robustness
- The persistence of a certain phenotypic trait or characteristic in a biological system despite perturbations or conditions of uncertainty. Robustness is achieved through the combination of many genetic and molecular mechanisms which effectively preserve the integrity of a particular adaptation, and can evolve by direct or indirect selection.
- runaway selection
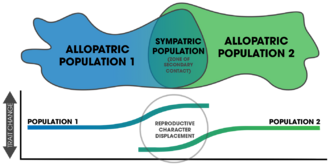
Reproductive character displacement sometimes occurs when two allopatric populations come into secondary contact. Once in sympatry, changes can be seen in mating-associated traits only in the zone of contact. This is a common pattern found in speciation by reinforcement.
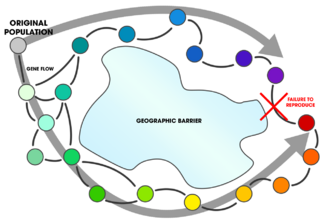
In a ring species, individuals are able to successfully reproduce and exchange genes with members of their own species in adjacent populations occupying a suitable habitat around a geographic barrier. Individuals at the ends of the cline are unable to reproduce when they come into contact.
S
- saltation
- A sudden and large mutational change from one generation to the next which is sufficient to cause rapid or immediate speciation. Various forms of saltation, such as by polyploidy in plants, have often historically been interpreted as evidence for certain theories of mutationism, in contrast to Darwinian gradualism.
- secondary contact
- The process by which two allopatrically distributed populations of a species are geographically reunited. Contact between divergent populations may renew the potential for gene flow between them, depending upon how reproductively isolated the populations have become.
- selection
- The non-random differential survival or reproduction of classes of phenotypically different entities.[1] Selection may occur naturally or may be induced artificially. Selection is often studied in different modes (as with sexual selection and kin selection) or from the perspective of distinct units (as with genic selection and group selection).
- selection coefficient
- The difference between the mean relative fitness of individuals of a given genotype and those of a reference genotype.[1]
- selective pressure
- selective sweep
- The process by which strong positive selection of a new and beneficial mutation within a population causes the mutation to reach fixation so quickly that nearby linked DNA sequences also become fixed via genetic hitchhiking, thereby reducing or eliminating the genetic variation of nearby loci within the population.
- semelparity
- A reproductive strategy characterized by a single reproductive episode during an individual organism's lifetime, especially one in which the programmed death of the organism immediately after the reproductive event constitutes part of an overall strategy that includes putting all available resources into maximizing the probability of reproductive success, at the expense of the organism's future life. Organisms that use such a strategy are said to be semelparous. Semelparity is usually contrasted with iteroparity.
- semi-geographic speciation
- semipermeable species boundary
- The idea that gene flow can occur between two species but that certain alleles at particular loci can exchange whereas others cannot.[21] It is often used to describe hybrid zones and has also been referred to as porous.[21]
- semispecies
- One of several groups of populations that are partially but not entirely reproductively isolated from each other by biological isolating mechanisms,[1] and which are therefore neither easily definable as belonging to the same species nor to separate species. The taxon of species itself is not a well-defined concept.
- sexual reproduction
- sexual selection
- spandrel
- speciation
- The evolutionary process by which populations evolve to become distinct species.
- speciation experiment
- An experiment that attempts to replicate reproductive isolation in nature in a scientifically controlled, laboratory setting.
- speciation in the fossil record
- Speciation that can be detected as occurring in fossilized organisms.
- speciation rate
- species
- A basic unit of biological classification, traditionally interpreted according to the biological species concept as the members in aggregate of a group of populations of organisms which interbreed or potentially interbreed with each other under natural conditions;[1] a basic taxonomic rank to which individual specimens are assigned and which often but not always corresponds to the definition of a biological species; and a fundamental unit used to interpret and measure biodiversity in ecological contexts. The concept of species is notoriously complex and often problematic to define precisely; many different conceptualizations of what is or should be meant by the term have been defined in scientific literature.
- species complex
- species concept
- species problem
- The difficulty in precisely defining what a species is and in determining the placement of an organism within a particular species.[31]
- stasipatric speciation
- stasis
- A species lineage that experiences little phenotypic or genotypic change over time.[7]
- stepping-stone speciation
- sterility
- subspecies
- A named geographic race, or a set of populations of the same species which share one or more distinctive features and occupy an area that is geographically separate from other subspecies.[1] Not all species are formally divided into subspecies, and the taxon of species itself is not a well-defined concept.
- survival of the fittest
- suture zone
- A geographic region that exhibits a significant number of hybrid zones, contact zones between populations, and phylogeographic breaks.[32]
- swamping effect
- sympatric speciation
- sympatry
- symplesiomorphy
- synapomorphy
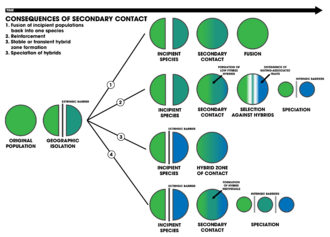
The four outcomes of secondary contact:
1. An extrinsic barrier separates a species population into two but they come into contact before reproductive isolation is sufficient to result in speciation. The two populations fuse back into one species.
2. Speciation by reinforcement.
3. Two separated populations stay genetically distinct while hybrid swarms form in the zone of contact.
4. Genome recombination results in speciation of the two populations, with an additional hybrid species. All three species are separated by intrinsic reproductive barriers.[30]
1. An extrinsic barrier separates a species population into two but they come into contact before reproductive isolation is sufficient to result in speciation. The two populations fuse back into one species.
2. Speciation by reinforcement.
3. Two separated populations stay genetically distinct while hybrid swarms form in the zone of contact.
4. Genome recombination results in speciation of the two populations, with an additional hybrid species. All three species are separated by intrinsic reproductive barriers.[30]
T
- taxon
- taxonomy
- teleonomy
- temporal isolation
- tension zone
- type
- type species
U
V
- vicariance biogeography
- A biogeographic approach to species distributions that uses their phylogenetic histories—patterns resulting from allopatric speciation events in the past.[33]
- vicariant speciation
- A biogeographic term meaning the geographic isolation of two species populations (as in allopatric speciation).
W
- Wahlund effect
- A phenomenon by which a reduction of heterozygosity at a particular genetic locus within a population as a whole is observed when two or more subpopulations have different allele frequencies at that locus, even if the subpopulations themselves are each in Hardy–Weinberg equilibrium.
- Wallace effect
Y
Z
See also
- Outline of evolution
- Index of evolutionary biology articles
- Glossary of cellular and molecular biology
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 1.25 1.26 Futuyma, Douglas J. (1986). Evolutionary biology (2nd ed.). Sunderland, Mass.: Sinauer Associates, Inc.. pp. 550–556. ISBN 0878931880. https://archive.org/stream/evolutionarybiol00futu#page/552/mode/2up. Retrieved 15 January 2021.
- ↑ Oxford Reference (2008), agamospecies, Oxford University Press
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Jerry A. Coyne; H. Allen Orr (2004), Speciation, Sinauer Associates, pp. 1–545, ISBN 978-0-87893-091-3
- ↑ Guy L. Bush (1994), "Sympatric speciation in animals: new wine in old bottles", Trends in Ecology & Evolution 9 (8): 285–288, doi:10.1016/0169-5347(94)90031-0, PMID 21236856
- ↑ Howard, Daniel J. (2003). "Speciation: Allopatric". Encyclopedia of Life Sciences. doi:10.1038/npg.els.0001748. ISBN 978-0470016176.
- ↑ Guy L. Bush (1994), "Sympatric speciation in animals: new wine in old bottles", Trends in Ecology & Evolution 9 (8): 285–288, doi:10.1016/0169-5347(94)90031-0, PMID 21236856
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 Vaux, Felix; Trewick, Steven A.; Morgan-Richards, Mary (2016). "Lineages, splits and divergence challenge whether the terms anagenesis and cladogenesis are necessary". Biological Journal of the Linnean Society 117 (2): 165–76. doi:10.1111/bij.12665.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 King, Robert C.; Stansfield, William D.; Mulligan, Pamela K. (2006). A Dictionary of Genetics (7th ed.). Oxford: Oxford University Press. ISBN 978-0-19-530762-7. https://openlibrary.org/works/OL2942141W/A_dictionary_of_genetics.
- ↑ Brian K. Hall (1984), "Developmental mechanisms underlying the atavisms", Biological Reviews 59 (1): 89–124, doi:10.1111/j.1469-185x.1984.tb00402.x, PMID 6367843
- ↑ M. V. Lomolino & J. H. Brown (1998), Biography (2 ed.), Sinauer Associates, Sunderland, MA., pp. 3, ISBN 978-0-87893-073-9, https://archive.org/details/biogeography0000brow/page/3
- ↑ Sergey Gavrilets (2000), "Patterns of Parapatric Speciation", Evolution 54 (4): 1126–1134, doi:10.1554/0014-3820(2000)054[1126:pops2.0.co;2], PMID 11005282
- ↑ Owen, Elizabeth; Daintith, Eve, eds (2004). The Facts on File Dictionary of Evolutionary Biology. New York: Market House Books Ltd. ISBN 0-8160-4924-6. https://books.google.com/books?id=9GXdGiZyzNAC.
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 13.6 Rieger, Rigomar (1991). Glossary of Genetics: Classical and Molecular (5th ed.). Berlin: Springer-Verlag. ISBN 3540520546. https://openlibrary.org/works/OL4093818W/Glossary_of_genetics?edition=glossaryofgeneti0000rieg.
- ↑ Page, Roderick DM. (2006). "Cospeciation". Encyclopedia of Life Sciences. doi:10.1038/npg.els.0004124. ISBN 978-0470016176.
- ↑ Howard D. Rundle and Patrik Nosil (2005), "Ecological Speciation", Ecology Letters 8 (3): 336–352, doi:10.1111/j.1461-0248.2004.00715.x
- ↑ "Founder takes all: density-dependent processes structure biodiversity". Trends in Ecology & Evolution 28 (2): 78–85. 2013. doi:10.1016/j.tree.2012.08.024. PMID 23000431.
- ↑ "A new definition of Genetic Counseling: National Society of Genetic Counselors' Task Force report". Journal of Genetic Counseling 15 (2): 77–83. April 2006. doi:10.1007/s10897-005-9014-3. PMID 16761103.
- ↑ Turelli, M; Orr, H.A. (May 1995). "The Dominance Theory of Haldane's Rule". Genetics 140 (1): 389–402. doi:10.1093/genetics/140.1.389. PMID 7635302.
- ↑ Anders Ödeen & Ann-Britt Florin (2002), "Sexual selection and peripatric speciation: the Kaneshiro model revisited", Journal of Evolutionary Biology 15 (2): 301–306, doi:10.1046/j.1420-9101.2002.00378.x
- ↑ B. M. Fitzpatrick; A. A. Fordyce; S. Gavrilets (2008), "What, if anything, is sympatric speciation?", Journal of Evolutionary Biology 21 (6): 1452–1459, doi:10.1111/j.1420-9101.2008.01611.x, PMID 18823452
- ↑ 21.0 21.1 21.2 21.3 21.4 21.5 Richard G. Harrison (2012), "The Language of Speciation", Evolution 66 (12): 3643–3657, doi:10.1111/j.1558-5646.2012.01785.x, PMID 23206125
- ↑ B. B. Fitzpatrick, J. A. Fordyce, & S. Gavrilets (2009), "Pattern, process and geographic modes of speciation", Journal of Evolutionary Biology 22 (11): 2342–2347, doi:10.1111/j.1420-9101.2009.01833.x, PMID 19732257
- ↑ 23.00 23.01 23.02 23.03 23.04 23.05 23.06 23.07 23.08 23.09 23.10 23.11 23.12 23.13 23.14 23.15 A Dictionary of Ecology, Evolution, and Systematics. New York: Cambridge University Press. 1982. ISBN 9780521239578.
- ↑ Childs, G.; Maxson, R.; Cohn, R. H.; Kedes, L. (1981). "Orphons: Dispersed genetic elements derived from tandem repetitive genes of eucaryotes". Cell 23 (3): 651–663. doi:10.1016/0092-8674(81)90428-1. PMID 6784929.
- ↑ Michael Turelli, Nicholas H. Barton, and Jerry A. Coyne (2001), "Theory and speciation", Trends in Ecology & Evolution 16 (7): 330–343, doi:10.1016/s0169-5347(01)02177-2, PMID 11403865
- ↑ Verne Grant (1971), Plant Speciation, New York: Columbia University Press, pp. 432, ISBN 978-0231083263
- ↑ Douglas J. Futuyma (1989), "Speciational trends and the role of species in macroevolution", The American Naturalist 134 (2): 318–321, doi:10.1086/284983
- ↑ Loren H. Rieseberg (2001), "Chromosomal rearrangements and speciation", Trends in Ecology & Evolution 16 (7): 351–358, doi:10.1016/s0169-5347(01)02187-5, PMID 11403867
- ↑ L. D. Gottlieb (2003), "Rethinking classic examples of recent speciation in plants", New Phytologist 161: 71–82, doi:10.1046/j.1469-8137.2003.00922.x
- ↑ Hvala, John A.; Wood, Troy E. (2012). Speciation: Introduction. doi:10.1002/9780470015902.a0001709.pub3. ISBN 978-0470016176.
- ↑ William P. Hanage (2013), "Fuzzy species revisited", BMC Biology 11 (41): 41, doi:10.1186/1741-7007-11-41, PMID 23587266
- ↑ Luciano Nicolas Naka, Catherine L. Bechtoldt, L. Magalli Pinto Henriques, and Robb T. Brumfield (2012), "The Role of Physical Barriers in the Location of Avian Suture Zones in the Guiana Shield, Northern Amazonia", The American Naturalist 179 (4): E115–E132, doi:10.1086/664627, PMID 22437185
- ↑ M. V. Lomolino & J. H. Brown (1998), Biography (2 ed.), Sinauer Associates, Sunderland, MA., pp. 352–357, ISBN 978-0-87893-073-9, https://archive.org/details/biogeography0000brow/page/352
 |
Licensed under CC BY-SA 3.0 | Source: https://handwiki.org/wiki/Biology:Glossary_of_evolutionary_biology14 views | Status: cached on August 26 2024 07:59:12↧ Download this article as ZWI file
 KSF
KSF