Haplogroup R1
Topic: Biology
 From HandWiki - Reading time: 9 min
From HandWiki - Reading time: 9 min
| Haplogroup R1 | |
|---|---|
 | |
| Possible place of origin | Siberia, Central Asia, South Asia or Southwest Asia[1][2][3] |
| Ancestor | R (R-M207) |
| Descendants | R1a (M420), R1b (M343) |
| Defining mutations | M173/P241/Page29, CTS916/M611/PF5859, CTS997/M612/PF6111, CTS1913/M654, CTS2565/M663, CTS2680, CTS2908/M666/PF6123, CTS3123/M670, CTS3321/M673, CTS4075/M682, CTS5611/M694, CTS7085/M716/Y481, CTS8116/M730, F93/M621/PF6114, F102/M625/PF6116, F132/M632, F211/Y290, F245/M659/Y477, FGC189/Y305, L875/M706/PF6131/YSC0000288, L1352/M785/YSC0000230, M306/PF6147/S1, M640/PF6118, M643, M689, M691/CTS4862/PF6042/YSC0001281, M710/PF6132/YSC0000192, M748/YSC0000207, M781, P225, P231, P233, P234, P236, P238/PF6115, P242/PF6113, P245/PF6117, P286/PF6136, P294/PF6112, PF6120[4] |
Haplogroup R1, or R-M173, is a Y-chromosome DNA haplogroup. A primary subclade of Haplogroup R (R-M207), it is defined by the SNP M173. The other primary subclade of Haplogroup R is Haplogroup R2 (R-M479).
Males carrying R-M173 in modern populations appear to comprise two subclades: R1a and R1b, which are found mainly in populations native to Eurasia (except East and Southeast Asia). R-M173 contains the majority of representatives of haplogroup R in the form of its subclades, R1a and R1b (Rosser 2000, Semino 2000).
Structure
| Haplogroup R1 | |||||||||||||||||||||||||||||||||||||||
|
Origins
R1 and its sibling clade R2 (R-M79) are the only immediate descendants of Haplogroup R (R-M207). R is a direct descendant of Haplogroup P1 (P-M45), and a sibling clade, therefore, of Haplogroup Q (Q-M242). The origins of haplogroup R1 cannot currently be proved. According to the SNP-Tracker (as of May 2023) it evolved around 25 000 BP/23 000 BC in western Siberia between the southern Urals and Lake Balkhash.[5]
No examples of the basal subclade, R1* have yet been identified in living individuals or ancient remains. However, the parent clade, R* was present in Upper Paleolithic-era individuals (24,000 years BP), from the Mal'ta-Buret' culture, in Siberia.[6] The autosomal DNA of the Mal'ta-Buret' people is a part of a group known to scholars of population genetics as Ancient North Eurasians (ANE). The first major descendant haplogroups appeared subsequently in hunter-gatherers from Eastern Europe (R1a, 13 kya)[7] and Western Europe (R1b, 14 kya),[8] with genotypes derived, to varying degrees, from ANE.[9]
General distribution
Eurasia
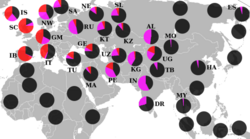
Haplogroup R1 is very common throughout all of Eurasia except East Asia and Southeast Asia. Its distribution is believed to be associated with the re-settlement of Eurasia following the Last Glacial Maximum. Its main subgroups are R1a and R1b. One subclade of haplogroup R1b (especially R1b1a2), is the most common haplogroup in Western Europe and Bashkortostan (Lobov 2009), while a subclade of haplogroup R1a (especially haplogroup R1a1) is the most common haplogroup in large parts of South Asia, Eastern Europe, Central Asia, Western China, and South Siberia.[10]
Individuals whose Y-chromosomes possess all the mutations on internal nodes of the Y-DNA tree down to and including M207 (which defines Haplogroup R) but which display neither the M173 mutation that defines haplogroup R1 nor the M479 mutation that defines Haplogroup R2 are categorized as belonging to group R* (R-M207). R* has been found in 10.3% (10/97) of a sample of Burusho and 6.8% (3/44) of a sample of Kalash from northern Pakistan (Firasat 2007).
Americas
The presence of haplogroup R1 among Indigenous Americans groups is a matter of controversy. It is now the most common haplogroup after the various Q-M242, especially in North America in Ojibwe people at 79%, Chipewyan 62%, Seminole 50%, Cherokee 47%, Dogrib 40% and Tohono O'odham 38%.
Some authorities point to the greater similarity between haplogroup R1 subclades found in North America and those found in Siberia (e.g. Lell [11] and Raghavan [12]), suggesting prehistoric immigration from Asia and/or Beringia.
Africa
One subclade, now known as R1b1a2 (R-V88), is found only at high frequencies amongst populations native to West Africa, such as the Fulani, and is believed to reflect a prehistoric back-migration from Eurasia to Africa.[citation needed]
Subclade distribution
R1a (R-M420)
The split of R1a (M420) is computed to ca 25,000 years ago (95% CI: 21, 300–29, 000 BP), or roughly the last glacial maximum. A large study performed in 2014 (Underhill et al. 2015), using 16,244 individuals from over 126 populations from across Eurasia, concluded that there was compelling evidence that "the initial episodes of haplogroup R1a diversification likely occurred in the vicinity of present-day Iran."[13] The subclade M417 (R1a1a1) diversified ca. 5,800 years ago.[14] The distribution of M417-subclades R1-Z282 (including R1-Z280)[15] in Central- and Eastern Europe and R1-Z93 in Asia[15][16] suggests that R1a1a diversified within the Eurasian Steppes or the Middle East and Caucasus region.[15] The place of origin of these subclades plays a role in the debate about the origins of the Indo-Europeans. High frequencies of haplogroup R1a are found amongst West Bengal Brahmins (72%), and Uttar Pradesh Brahmins, (67%), the Ishkashimi (68%), the Tajik population of Panjikent (64%), the Kyrgyz population of Central Kyrgyzstan (63.5%), Sorbs (63.39%), Bihar Brahmins (60.53%), Shors (58.8%),[17] Poles (56.4%), Teleuts (55.3%),[17] South Altaians (58.1%),[18] Ukrainians (50%) and Russians (50%) (Semino 2000, Wells 2001, Behar 2003, and Sharma 2007).
R1b (R-M343)
Haplogroup R1b probably originated in Eurasia prior to or during the last glaciation. It is the most common haplogroup in Western Europe and Bashkortostan.(Lobov 2009) It may have survived the last glacial maximum,[19] in refugia near the southern Ural Mountains and Aegean Sea.(Lobov 2009).
It is also present at lower frequencies throughout Eastern Europe, with higher diversity than in western Europe, suggesting an ancient migration of haplogroup R1b from the east.[20] Haplogroup R1b is also found at various frequencies in many different populations near the Ural Mountains and Central Asia, its likely region of origin.
There may be a correlation between this haplogroup and the spread of Centum branch Indo-European languages in southern and western Europe. For instance, the modern incidence of R1b reaches between 60% and 90% of the male population in most parts of Spain , Portugal, France , Britain and Ireland.[21] The clade is also found at frequencies of up to 90% in the Chad Basin, and is also present in North Africa, where its frequency surpasses 10% in some parts of Algeria.
Although it is rare in South Asia, some populations show relatively high percentages for R1b. These include Lambadi showing 37%(Kivisild 2003). Hazara 32% (Sengupta 2005), and Agharia (in East India) at 30% (Sengupta 2005). Besides these, R1b has appeared in Balochi (8%), Bengalis (6.5%), Chenchu (2%), Makrani (5%), Newars (10.6%), Pallan (3.5%) and Punjabis (7.6%) (Kivisild 2003, Sengupta 2005, and Gayden 2007). In Southeast Asia, it is present in the Philippines due to Spanish and American colonization where different studies vary as to its frequency; from 3.6% of the male population, in a year 2001 study conducted by Stanford University Asia-Pacific Research Center had European Y-DNA R1b to 13% in an Public Y-DNA Library.[22][23][24]
R-M343 (previously called Hg1[citation needed] and Eu18[citation needed]) is the most frequent Y-chromosome haplogroup in Europe. It is an offshoot of R-M173, characterised by the M343 marker.[25] An overwhelming majority of members of R-M343 are classified as R-P25 (defined by the P25 marker), the remainder as R-M343*. Its frequency is highest in Western Europe (and due to modern European immigration, in parts of the Americas). The majority of R-M343-carriers of European descent belong to the R-M269 (R1b1a2) descendant line.
See also
Genetics
Y-DNA R-M207 subclades
References
- ↑ Kivisild 2003
- ↑ Soares 2010
- ↑ (Wells 2001) [|permanent dead link|dead link}}]
- ↑ Y-DNA Haplogroup R and its Subclades – 2008 from ISOGG
- ↑ "SNP Tracker". http://scaledinnovation.com/gg/snpTracker.html.
- ↑ Haak, Wolfgang; Lazaridis, Iosif; Patterson, Nick; Rohland, Nadin; Mallick, Swapan; Llamas, Bastien; Brandt, Guido; Nordenfelt, Susanne et al. (10 February 2015). "Massive migration from the steppe is a source for Indo-European languages in Europe" (in en). bioRxiv: 013433. doi:10.1101/013433. http://biorxiv.org/lookup/doi/10.1101/013433.
- ↑ Saag, Lehti; Vasilyev, Sergey V.; Varul, Liivi; Kosorukova, Natalia V.; Gerasimov, Dmitri V.; Oshibkina, Svetlana V.; Griffith, Samuel J.; Solnik, Anu et al. (January 2021). "Genetic ancestry changes in Stone to Bronze Age transition in the East European plain" (in en). Science Advances 7 (4): eabd6535. doi:10.1126/sciadv.abd6535. PMID 33523926. Bibcode: 2021SciA....7.6535S.
- ↑ "The genetic history of Ice Age Europe". Nature 534 (7606): 200–5. June 2016. doi:10.1038/nature17993. PMID 27135931. Bibcode: 2016Natur.534..200F.
- ↑ Haak, Wolfgang; Lazaridis, Iosif; Patterson, Nick; Rohland, Nadin; Mallick, Swapan; Llamas, Bastien; Brandt, Guido; Nordenfelt, Susanne et al. (June 2015). "Massive migration from the steppe was a source for Indo-European languages in Europe" (in en). Nature 522 (7555): 207–211. doi:10.1038/nature14317. ISSN 1476-4687. PMID 25731166. Bibcode: 2015Natur.522..207H.
- ↑ "Results for R1b1 members". http://homepage.eircom.net/~ihdp/ihdp/r1b1c.htm.
- ↑ Lell Jeffrey T.; Sukernik Rem I.; Starikovskaya Yelena B.; Su Bing; Jin Li; Schurr Theodore G.; Underhill Peter A.; Wallace Douglas C. (2002). "The Dual Origin and Siberian Affinities of Native American". The American Journal of Human Genetics 70 (1): 192–206. doi:10.1086/338457. PMID 11731934.
- ↑ Raghavan Maanasa et al. (2013). "(2 January 2014). "Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans"". Nature 505 (7481): 87–91. doi:10.1038/nature12736. PMID 24256729. Bibcode: 2014Natur.505...87R.
- ↑ Underhill, Peter A. (2015), "The phylogenetic and geographic structure of Y-chromosome haplogroup R1a", European Journal of Human Genetics 23 (1): 124–131, doi:10.1038/ejhg.2014.50, PMID 24667786
- ↑ Underhill 2014, p. 130.
- ↑ 15.0 15.1 15.2 Pamjav 2012.
- ↑ Underhill 2014.
- ↑ 17.0 17.1 Miroslava Derenko et al 2005, Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions
- ↑ Khar'kov, V.N. (2007), "Gene pool differences between Northern and Southern Altaians inferred from the data on Y-chromosomal haplogroups", Genetika 43 (5): 675–87, doi:10.1134/S1022795407050110, PMID 17633562
- ↑ "Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample". Int. J. Legal Med. 122 (3): 251–5. May 2008. doi:10.1007/s00414-007-0203-5. PMID 17909833.
- ↑ "Variations of R1b Ydna in Europe: Distribution and Origins | WorldFamilies.net". https://www.worldfamilies.net/Tools/r1b_ydna_in_europe.
- ↑ Most Euro men are related to King Tut: DNA testing reveals strange genetic link among Europeans; Oddly, most Egyptians not in the family, Metro NY, 2 August 2011, http://www.metro.us/newyork/international/article/931992--most-euro-men-are-related-to-king-tut, retrieved 14 September 2011
- ↑ "With a sample population of 105 Filipinos, the company of Applied Biosystems, analysed the Y-DNA of average Filipinos and it is discovered that about 0.95% of the samples have the Y-DNA Haplotype "H1a", which is most common in South Asia and had spread to the Philippines via precolonial Indian missionaries who spread Hinduism and established Indic Rajahnates like Cebu and Butuan. The 13% frequeny of R1b also indicate Spanish admixture". http://www6.appliedbiosystems.com/yfilerdatabase/.
- ↑ "Manual Collation". https://docs.google.com/spreadsheets/d/1wD-JtgTGL3qQaDcZi8CSPZaBSkqXEIKkoaWiBR1bgp8/edit#gid=0.
- ↑ Philippines DNA Project - Y-DNA Classic Chart
- ↑ Note that in earlier literature the M269 marker, rather than M343, was used to define the "R1b" haplogroup. Then, for a time (from 2003 to 2005) what is now R1b1c was designated R1b3.
Works cited
- Gayden, T; Cadenas, AM; Regueiro, M; Singh, NB; Zhivotovsky, LA; Underhill, PA; Cavalli-Sforza, LL; Herrera, RJ (2007), "The Himalayas as a directional barrier to gene flow.", American Journal of Human Genetics 80 (5): 884–94, doi:10.1086/516757, PMID 17436243
- Behar; Thomas, MG; Skorecki, K; Hammer, MF; Bulygina, E; Rosengarten, D; Jones, AL; Held, K et al. (2003), "Multiple Origins of Ashkenazi Levites: Y Chromosome Evidence for Both Near Eastern and European Ancestries", Am. J. Hum. Genet. 73 (4): 768–779, doi:10.1086/378506, PMID 13680527, PMC 1180600, http://www.ucl.ac.uk/tcga/tcgapdf/Behar-AJHG-03.pdf
- Luigi Luca Cavalli-Sforza (1994), The History and Geography of Human Genes, Princeton University Press, ISBN 978-0-691-08750-4
- Cinnioğlu, C et al. (2004), "Excavating Y-chromosome haplotype strata in Anatolia", Hum Genet 114 (2): 127–48, doi:10.1007/s00439-003-1031-4, PMID 14586639
- Firasat, S.; Khaliq, S.; Mohyuddin, A.; Papaioannou, M.; Tyler-Smith, C.; Underhill, P. A.; Ayub, Q. (2007). "Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan". European Journal of Human Genetics 15 (1): 121–26. doi:10.1038/sj.ejhg.5201726. PMID 17047675.
- Kivisild, T.; Rootsi, S.; Metspalu, M.; Mastana, S.; Kaldma, K.; Parik, J.; Metspalu, E.; Adojaan, M. et al. (2003). "The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations". American Journal of Human Genetics 72 (2): 313–332. doi:10.1086/346068. ISSN 0002-9297. PMID 12536373.
- "Structure of the Gene Pool of Bashkir Subpopulations" (in ru). 2009. http://ftp.anrb.ru/molgen/Lobov_AS.PDF.
- Pamjav (December 2012), "Brief communication: New Y-chromosome binary markers improve phylogenetic resolution within haplogroup R1a1", American Journal of Physical Anthropology 149 (4): 611–615, doi:10.1002/ajpa.22167, PMID 23115110
- Passarino et al. (2002), "Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms", Eur. J. Hum. Genet. 10 (9): 521–9, doi:10.1038/sj.ejhg.5200834, PMID 12173029
- Saha et al. (2005), "Genetic affinity among five different population groups in India reflecting a Y-chromosome gene flow", J. Hum. Genet. 50 (1): 49–51, doi:10.1007/s10038-004-0219-3, PMID 15611834
- Semino (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans", Science 290 (5494): 1155–9, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S, http://hpgl.stanford.edu/publications/Science_2000_v290_p1155.pdf
- Sengupta et al. (2005), "Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists", Am. J. Hum. Genet. 78 (2): 202–21, doi:10.1086/499411, PMID 16400607
- Sharma, Swarkar; Rai, Ekta; Bhat, Audesh K; Bhanwer, Amarjit S; Bamezai, Rameshwar NK (2007). "A novel subgroup Q5 of human Y-chromosomal haplogroup Q in India". BMC Evolutionary Biology 7 (1): 232. doi:10.1186/1471-2148-7-232. PMID 18021436. Bibcode: 2007BMCEE...7..232S.
- Soares et al. (2010), "The Archaeogenetics of Europe", Current Biology 20 (4): R174–83, doi:10.1016/j.cub.2009.11.054, PMID 20178764
- Wells et al. (2001), "The Eurasian Heartland: A continental perspective on Y-chromosome diversity", Proc. Natl. Acad. Sci. U.S.A. 98 (18): 10244–9, doi:10.1073/pnas.171305098, PMID 11526236, Bibcode: 2001PNAS...9810244W. Also [1]
- "Y-Chromosomal Diversity in Europe Is Clinal and Influenced Primarily by Geography, Rather than by Language". American Journal of Human Genetics 67 (6): 1526–1543. 2000. doi:10.1086/316890. PMID 11078479.
 |
 KSF
KSF