Hopanoids
Topic: Biology
 From HandWiki - Reading time: 14 min
From HandWiki - Reading time: 14 min

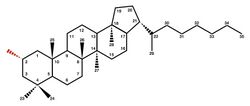
Hopanoids are a diverse subclass of triterpenoids with the same hydrocarbon skeleton as the compound hopane. This group of pentacyclic molecules therefore refers to simple hopenes, hopanols and hopanes, but also to extensively functionalized derivatives such as bacteriohopanepolyols (BHPs) and hopanoids covalently attached to lipid A.[1][2]
The first known hopanoid, hydroxyhopanone, was isolated by two chemists at The National Gallery, London working on the chemistry of dammar gum, a natural resin used as a varnish for paintings.[3] While hopanoids are often assumed to be made only in bacteria, their name actually comes from the abundance of hopanoid compounds in the resin of plants from the genus Hopea. In turn, this genus is named after John Hope, the first Regius Keeper of the Royal Botanic Garden, Edinburgh.
Since their initial discovery in an angiosperm, hopanoids have been found in plasma membranes of bacteria, lichens, bryophytes, ferns, tropical trees and fungi.[4] Hopanoids have stable polycyclic structures that are well-preserved in petroleum reservoirs, rocks and sediment, allowing the diagenetic products of these molecules to be interpreted as biomarkers for the presence of specific microbes and potentially for chemical or physical conditions at the time of deposition.[5] Hopanoids have not been detected in archaea.[6][7]
Biological function
About 10% of sequenced bacterial genomes have a putative shc gene encoding a squalene-hopene cyclase and can presumably make hopanoids, which have been shown to play diverse roles in the plasma membrane and may allow some organisms to adapt in extreme environments.[8][9]
Since hopanoids modify plasma membrane properties in bacteria, they are frequently compared to sterols (e.g., cholesterol), which modulate membrane fluidity and serve other functions in eukaryotes.[10] Although hopanoids do not rescue sterol deficiency, they are thought to increase membrane rigidity and decrease permeability.[9][11][12] Also, gammaproteobacteria and eukaryotic organisms such as lichens and bryophytes have been shown to produce both sterols and hopanoids, suggesting these lipids may have other distinct functions.[4][13] Notably, the way hopanoids pack into the plasma membrane can change depending on what functional groups are attached. The hopanoid bacteriohopanetetrol assumes a transverse orientation in lipid bilayers, but diploptene localizes between the inner and outer leaflet, presumably thickening the membrane to decrease permeability.[14]
The hopanoid diplopterol orders membranes by interacting with lipid A, a common membrane lipid in bacteria, in ways similar to how cholesterol and sphingolipids interact in eukaryotic plasma membranes.[10] Diplopterol and cholesterol were demonstrated to promote condensation and inhibit gel-phase formation in both sphingomyelin monolayers and monolayers of glycan-modified lipid A. Furthermore, both diplopterol and cholesterol could rescue pH-dependent phase transitions in glycan-modified lipid A monolayers.[10] The role of hopanoids in membrane-mediated acid tolerance is further supported by observations of acid-inhibited growth and morphological abnormalities of the plasma membrane in hopanoid-deficient bacteria with mutant squalene-hopene cyclases.[15][16]
Hopanoids are produced in several nitrogen-fixing bacteria.[9] In the actinomycete Frankia, hopanoids in the membranes of vesicles specialized for nitrogen fixation likely restrict the entry of oxygen by making the lipid bilayer more tight and compact.[17] In Bradyrhizobium, hopanoids chemically bonded to lipid A increase membrane stability and rigidity, enhancing stress tolerance and intracellular survival in Aeschynomene legumes.[18] In the cyanobacterium Nostoc punctiforme, large quantities of 2-methylhopanoids localize to the outer membranes of survival structures called akinetes.[19] In another example of stress tolerance, hopanoids in the aerial hyphae (spore bearing structures) of the prokaryotic soil bacteria Streptomyces are thought to minimize water loss across the membrane to the air.[20]
Biosynthesis
Squalene synthesis
Since hopanoids are a C30 terpenoid, biosynthesis begins with isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAP), which are combined to form longer chain isoprenoids.[2] Synthesis of these smaller precursors proceeds either via the mevalonate pathway or the methylerythritol-4-phosphate pathway depending on the bacterial species, although the latter tends to be more common.[21] DMAP condenses with one molecule of IPP to geranyl pyrophosphate, which in turn condenses with another IPP to generate farnesyl pyrophosphate (FPP).[2] Squalene synthase, coded for by the gene sqs, then catalyzes the condensation of two FPP molecules to presqualene pyrophosphate (PSPP) before oxidizing NADPH to release squalene.[22] However, some hopanoid-producing bacteria lack squalene synthase and instead use the three enzymes HpnC, HpnD and HpnE, which are encoded in the hpn operon with many other hopanoid biosynthesis genes.[23] In this alternative yet seemingly more widespread squalene synthesis pathway, HpnD releases pyrophosphate as it condenses two molecules of FPP to PSPP, which HpnC converts to hydroxysqualene, consuming a water molecule and releasing another pyrophosphate. Then, hydroxysqualene is reduced to squalene in a dehydration reaction mediated by the FAD-dependent enzyme HpnE.[22]
Cyclization
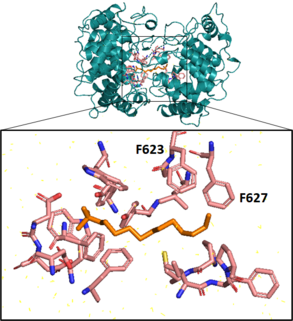
Next, a squalene-hopene cyclase catalyzes an elaborate cyclization reaction, engaging squalene in an energetically favorable all-chair conformation before creating 5 cycles, 6 covalent bonds, and 9 chiral centers on the molecule in a single step.[24][25] This enzyme, coded for by the gene shc (also called hpnF in some bacteria), has a double ⍺-barrel fold characteristic of terpenoid biosynthesis[26] and is present in the cell as a monotopic homodimer, meaning pairs of the cyclase are embedded in but do not span the plasma membrane.[24][27] In vitro, this enzyme exhibits promiscuous substrate specificity, also cyclizing 2,3-oxidosqualene.[28]
Aromatic residues in the active site form several unfavorable carbocations on the substrate which are quenched by a rapid polycyclization.[25] In the last substep of the cyclization reaction, after electrons comprising the terminal alkene bond on the squalene have attacked the hopenyl carbocation to close the E ring, the C22 carbocation may be quenched by mechanisms that lead to different hopanoid products. Nucleophilic attack of water will yield diplopterol, while deprotonation at an adjacent carbon will form one of several hopene isomers, often diploptene.[4]
Functionalization
After cyclization, hopanoids are frequently modified by hopanoid biosynthesis enzymes encoded by genes in the same operon as shc, hpn.[29] For instance, the radical SAM protein HpnH adds an adenosine group to diploptene, forming the extended C35 hopanoid adenosylhopane, which can then be further functionalized by other hpn gene products.[30] HpnG catalyzes the removal of adenine from adenosylhopane to make ribosyl hopane, which reacts to form bacteriohopanetetrol (BHT) in a reaction mediated by an unknown enzyme.[31] Additional modifications may occurs as HpnO aminates the terminal hydroxyl on BHT, producing amino bacteriohopanetriol, or as the glycosyltransferase HpnI converts BHT to N-acetylglucosaminyl-BHT.[32] In sequence, the hopanoid biosynthesis associated protein HpnK mediates deacetylation to glucosaminyl-BHT, from which radical SAM protein HpnJ generates a cyclitol ether.[32]
Importantly, C30 and C35 hopanoids alike may be methylated at C2 and C3 positions by the radical SAM methyltransferases HpnP and HpnR, respectively.[33][34] These two methylations are particularly geostable compared to side-chain modifications and have entertained geobiologists for decades.[9]
In a biosynthetic pathway exclusive to some bacteria, the enzyme tetrahymanol synthase catalyzes the conversion of the hopanoid diploptene to the pentacyclic triterpenoid tetrahymanol. In eukaryotes like Tetrahymena, tetrahymanol is instead synthesized directly from squalene by a cyclase with no homology to the bacterial tetrahymanol synthase.[35]
In paleobiology
Hopanoids have been estimated to be the most abundant natural products on Earth, remaining in the organic fraction of all sediments, independent of age, origin or nature. The total amount in the Earth was estimated as 10 x 1018 gram (1012 ton) in 1992.[36] Biomolecules like DNA and proteins are degraded during diagenesis, but polycyclic lipids persist in the environment over geologic timescales due to their fused, stable structures.[37] Although hopanoids and sterols are reduced to hopanes and steranes during deposition, these diagenetic products can still be useful biomarkers, or molecular fossils, for studying the coevolution of early life and Earth.[37][38]
Currently, the oldest detected undisputed triterpenoid fossils are Mesoproterozoic okenanes, steranes, and methylhopanes from a 1.64 Ga (billion year) old basin in Australia.[39] However, molecular clock analyses estimate that the earliest sterols were likely produced around 2.3 Ga ago, around the same time as the Great Oxidation Event, with hopanoid synthesis arising even earlier.[40]
For several reasons, hopanoids and squalene-hopene cyclases have been hypothesized to be more ancient than sterols and oxidosqualene cyclases. First, diplopterol is synthesized when water quenches the C22 carbocation formed during polycyclization. This indicates that hopanoids can be made without molecular oxygen and could have served as a sterol surrogate before the atmosphere accumulated oxygen, which reacts with squalene in a reaction catalyzed by squalene monooxygenase during sterol biosynthesis.[1] Furthermore, squalene binds to squalene-hopene cyclases in a low-energy, all-chair conformation while oxidosqualene is cyclized in a more strained, chair-boat-chair-boat conformation.[4][41] Squalene-hopene cyclases also display more substrate promiscuity in that they cyclize oxidosqualene in vitro, causing some scientists to hypothesize that they are evolutionary predecessors to oxidosqualene cyclases.[41] Other scientists have proposed that squalene-hopene and oxidosqualene cyclases diverged from a common ancestor, a putative bacterial cyclase that would have made a tricyclic malabaricanoid or tetracyclic dammaranoid product.[1][42]
2-methylhopanoids
As a biomarker for cyanobacteria
Proposal
2-methylhopanes, often quantified as the 2-α-methylhopane index, were first proposed as a biomarker for oxygenic photosynthesis by Roger Summons and colleagues following the discovery of the precursor lipids, 2-methylhopanols, in cyanobacterial cultures and mats.[43] The subsequent discovery of 2-α-methylhopanes supposedly from photosynthetic cyanobacteria in 2.7 Ga old shales from the Pilbara Craton of Western Australia suggested a 400 Ma (million year) gap between the evolution of oxygenic metabolism and the Great Oxidation Event.[44] However, these findings were later rejected due to potential contamination by modern hydrocarbons.[45]
Putative cyanobacterial presence on the basis of abundant 2-methylhopanes has been used to explain black shale deposition during Aptian and Cenomanian–Turonian Ocean Anoxic Events (OAEs) and the associated 15N isotopic signatures indicative of N2-fixation.[46] In contrast, 2-α-methylhopane index values are relatively low across similar Frasnian and Famennian sediments corresponding to the Kellwasser event(s),[47] though higher levels have been reported in later Lower Famennian sections.[48]
Dispute
The status of 2-methylhopanoids as a cyanobacterial biomarker was challenged by a number of microbiological discoveries. Geobacter sulfurreducens was demonstrated to synthesize diverse hopanols, although not 2-methyl-hopanols, when grown under strictly anaerobic conditions.[8] Furthermore, the anoxygenic phototroph Rhodopseudomonas palustris was found to produce 2-methyl-BHPs only under anoxic conditions.[49] This latter discovery also lead to the identification of the gene encoding the key methyltransferase HpnP.[33] hpnP was subsequently identified in an acidobacterium and numerous alphaproteobacteria, and phylogenetic analysis of the gene concluded that it originated in the alphaproteobacteria and was acquired by the cyanobacteria and acidobacteriota via horizontal gene transfer.[50]
Among cyanobacteria, hopanoid production is generally limited to terrestrial cyanobacteria. Among marine cyanobacteria, culture experiments in conducted by Helen Talbot and colleagues concluded that only two marine species–Trichodesmium and Crocosphaera–produced bacteriohopanepolyols.[51] A later gene-based search for hpnP in available cyanobacterial genomes and Metagenome Assembled Genomes (MAGs) drew similar conclusions, identifying the gene in ~30% of terrestrial and freshwater species, and only one of the 739 marine cyanobacterial genomes and MAGs.[52] Additionally, Nostoc punctiforme produces the greatest amount of 2-methylhopanoids when differentiated into akinetes. These cold- and desiccation-resistant cell structures are dormant and therefore not photosynthetically active, further challenging the association between 2-methylhopanes and oxygenic photosynthesis.[19]
Other interpretations
Research demonstrating that the nitrite-oxidizing bacteria (NOB) Nitrobacter vulgaris increases its production of 2-methylhopanoids 33-fold when supplemented with cobalamin has furthered a non-cyanobacterial explanation for the observed abundance of 2-methylhopanes associated with Cretaceous OAEs. Felix Elling and colleagues propose that overturning circulation brought ammonia- and cobalt-rich deep waters to the surface, promoting aerobic nitrite oxidation and cobalamin synthesis, respectively. This model also addresses the conspicuous lack of 2-methylhopanes associated with Mediterranean sapropel events and in modern Black Sea sediments. Because both environments feature much less upwelling, 2-methylhopanoid-producing NOB such as N. vulgaris are outcompeted by NOB with higher nitrite affinity and anammox bacteria.[52]
An environmental survey by Jessica Ricci and coauthors using metagenomes and clone libraries found significant correlation between plant-associated microbial communities and hpnP presence, based on which they propose that 2-methylhopanoids are a biomarker for sessile microbial communities high in osmolarity and low in oxygen and fixed nitrogen.[53]
3-methylhopanoids
3-methylhopanoids have historically been associated with aerobic methanotrophy based on culture experiments[54] and co-occurrence with aerobic methanotrophs in the environment.[55] As such, the presence of 3-methylhopanes, together with 13C depletion, are considered markers of ancient aerobic methanotrophy.[34] However, acetic acid bacteria have been known for decades to also produce 2-methylhopanoids.[54] Additionally, following their identification of hpnR, the gene responsible for methylating hopanoids at the C3 position, Paula Welander and Roger Summons identified putative hpnR homologs in members of alpha-, beta-, and gammaproteobacteria, actinomycetota, nitrospirota, candidate phylum NC10, and an acidobacterium, as well as in three metagenomes. As such, Welander and Summons conclude that 3-methylhopanoids alone cannot constitute evidence of aerobic methanotrophy.[34]
Applications
Industry
The elegant mechanism behind the protonase activity of squalene-hopene cyclase was appreciated and adapted by chemical engineers at the University of Stuttgart, Germany. Active site engineering resulted in loss of the enzyme's ability to form hopanoids, but enabled Brønsted acid catalysis for the stereoselective cyclization of the monoterpenoids geraniol, epoxygeraniol, and citronellal.[56]
Agriculture
The application of hopanoids and hopanoid-producing nitrogen fixers to soil has been proposed and patented as a biofertilizer technique that increases environmental resistance of plant-associated microbial symbionts, including nitrogen-fixing bacteria that are essential for transforming atmospheric nitrogen to soluble forms available to crops.[57]
Medicine
During later studies of interactions between diplopterol and lipid A in Methylobacterium extorquens, multidrug transport was found to be a hopanoid-dependent process. Squalene-hopene cyclase mutants derived from a wild type capable of multidrug efflux, a drug-resistance mechanism mediated by integral transport proteins, lost the ability to perform both multidrug transport and hopanoid synthesis.[12] Researchers indicate this could be due to direct regulation of transport proteins by hopanoids or indirectly by altering membrane ordering in a way that disrupts the transport system.[12]
References
- ↑ 1.0 1.1 1.2 "Deciphering the evolutionary history of microbial cyclic triterpenoids". Free Radical Biology & Medicine. Early Life on Earth and Oxidative Stress 140: 270–278. August 2019. doi:10.1016/j.freeradbiomed.2019.05.002. PMID 31071437.
- ↑ 2.0 2.1 2.2 "Bacterial membrane lipids: diversity in structures and pathways". FEMS Microbiology Reviews 40 (1): 133–59. January 2016. doi:10.1093/femsre/fuv008. PMID 25862689.
- ↑ "The chemistry of dammar resin" (in en). Journal of the Chemical Society (Resumed): 3132–3140. 1955-01-01. doi:10.1039/JR9550003132. ISSN 0368-1769.
- ↑ 4.0 4.1 4.2 4.3 "Sterols and other triterpenoids: source specificity and evolution of biosynthetic pathways" (in en). Organic Geochemistry 36 (2): 139–159. 2005-02-01. doi:10.1016/j.orggeochem.2004.06.013.
- ↑ "Early developments in petroleum geochemistry" (in en). Organic Geochemistry 33 (9): 1025–1052. 2002-09-01. doi:10.1016/S0146-6380(02)00056-6.
- ↑ William W. Christie. "The AOCS Lipid Library. Hopanoids". American Oil Chemists' Society. Archived from the original on 2016-03-05. Retrieved 2015-11-17.
- ↑ "Hopanoids - AOCS Lipid Library". 2016-03-05. http://lipidlibrary.aocs.org/Primer/content.cfm?ItemNumber=39334.
- ↑ 8.0 8.1 "Targeted genomic detection of biosynthetic pathways: anaerobic production of hopanoid biomarkers by a common sedimentary microbe". Geobiology 3 (1): 33–40. 2005. doi:10.1111/j.1472-4669.2005.00041.x.
- ↑ 9.0 9.1 9.2 9.3 "Hopanoid lipids: from membranes to plant-bacteria interactions". Nature Reviews. Microbiology 16 (5): 304–315. May 2018. doi:10.1038/nrmicro.2017.173. PMID 29456243.
- ↑ 10.0 10.1 10.2 "Functional convergence of hopanoids and sterols in membrane ordering". Proceedings of the National Academy of Sciences of the United States of America 109 (35): 14236–40. August 2012. doi:10.1073/pnas.1212141109. PMID 22893685. Bibcode: 2012PNAS..10914236S.
- ↑ "Methylation at the C-2 position of hopanoids increases rigidity in native bacterial membranes". eLife 4: e05663. January 2015. doi:10.7554/eLife.05663. PMID 25599566.
- ↑ 12.0 12.1 12.2 "Hopanoids as functional analogues of cholesterol in bacterial membranes". Proceedings of the National Academy of Sciences of the United States of America 112 (38): 11971–6. September 2015. doi:10.1073/pnas.1515607112. PMID 26351677. Bibcode: 2015PNAS..11211971S.
- ↑ "Squalene-hopene cyclase from Methylococcus capsulatus (Bath): a bacterium producing hopanoids and steroids". Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism 1391 (2): 223–32. March 1998. doi:10.1016/S0005-2760(97)00212-9. PMID 9555026.
- ↑ "The relative effect of sterols and hopanoids on lipid bilayers: when comparable is not identical". The Journal of Physical Chemistry B 117 (50): 16129–40. December 2013. doi:10.1021/jp409748d. PMID 24299489.
- ↑ "Hopanoid production is required for low-pH tolerance, antimicrobial resistance, and motility in Burkholderia cenocepacia". Journal of Bacteriology 193 (23): 6712–23. December 2011. doi:10.1128/JB.05979-11. PMID 21965564.
- ↑ "Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1". Journal of Bacteriology 191 (19): 6145–56. October 2009. doi:10.1128/JB.00460-09. PMID 19592593.
- ↑ "Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase". Proceedings of the National Academy of Sciences of the United States of America 90 (13): 6091–4. July 1993. doi:10.1073/pnas.90.13.6091. PMID 11607408. Bibcode: 1993PNAS...90.6091B.
- ↑ "Covalently linked hopanoid-lipid A improves outer-membrane resistance of a Bradyrhizobium symbiont of legumes". Nature Communications 5 (1): 5106. October 2014. doi:10.1038/ncomms6106. PMID 25355435. Bibcode: 2014NatCo...5.5106S.
- ↑ 19.0 19.1 "2-Methylhopanoids are maximally produced in akinetes of Nostoc punctiforme: geobiological implications". Geobiology 7 (5): 524–32. December 2009. doi:10.1111/j.1472-4669.2009.00217.x. PMID 19811542.
- ↑ "Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2)". FEMS Microbiology Letters 189 (1): 93–5. August 2000. doi:10.1111/j.1574-6968.2000.tb09212.x. PMID 10913872.
- ↑ "Metabolic plasticity for isoprenoid biosynthesis in bacteria". The Biochemical Journal 452 (1): 19–25. May 2013. doi:10.1042/BJ20121899. PMID 23614721. https://portlandpress.com/biochemj/article-abstract/452/1/19/46166/Metabolic-plasticity-for-isoprenoid-biosynthesis?redirectedFrom=fulltext.
- ↑ 22.0 22.1 "Biosynthesis of Squalene from Farnesyl Diphosphate in Bacteria: Three Steps Catalyzed by Three Enzymes". ACS Central Science 1 (2): 77–82. 2015-05-27. doi:10.1021/acscentsci.5b00115. PMID 26258173.
- ↑ "Bacteria Do It Differently: An Alternative Path to Squalene". ACS Central Science 1 (2): 64–5. May 2015. doi:10.1021/acscentsci.5b00142. PMID 27162951.
- ↑ 24.0 24.1 "Squalene-hopene cyclases". Applied and Environmental Microbiology 77 (12): 3905–15. June 2011. doi:10.1128/AEM.00300-11. PMID 21531832.
- ↑ 25.0 25.1 "Squalene-hopene cyclase: catalytic mechanism and substrate recognition". Chemical Communications (4): 291–301. February 2002. doi:10.1039/B108995C. PMID 12120044.
- ↑ "Squalene-hopene cyclases-evolution, dynamics and catalytic scope". Current Opinion in Structural Biology. Multi-protein assemblies in signaling • Catalysis and regulation 41: 73–82. December 2016. doi:10.1016/j.sbi.2016.05.019. PMID 27336183.
- ↑ "Site-directed mutagenesis of squalene-hopene cyclase: altered substrate specificity and product distribution" (in English). Chemistry & Biology 7 (8): 643–9. August 2000. doi:10.1016/S1074-5521(00)00003-X. PMID 11048954.
- ↑ "Non-specific biosynthesis of hopane triterpenes by a cell-free system from Acetobacter pasteurianum". European Journal of Biochemistry 112 (3): 541–7. December 1980. doi:10.1111/j.1432-1033.1980.tb06117.x. PMID 7460938.
- ↑ "Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids". Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism 1393 (1): 108–18. July 1998. doi:10.1016/S0005-2760(98)00064-2. PMID 9714766.
- ↑ "Adenosylhopane: The first intermediate in hopanoid side chain biosynthesis" (in en). Organic Geochemistry 41 (10): 1075–1081. 2010-10-01. doi:10.1016/j.orggeochem.2010.07.003.
- ↑ "Ribosylhopane, a novel bacterial hopanoid, as precursor of C35 bacteriohopanepolyols in Streptomyces coelicolor A3(2)". ChemBioChem 15 (14): 2156–61. September 2014. doi:10.1002/cbic.201402261. PMID 25155017.
- ↑ 32.0 32.1 "Elucidation of the Burkholderia cenocepacia hopanoid biosynthesis pathway uncovers functions for conserved proteins in hopanoid-producing bacteria". Environmental Microbiology 17 (3): 735–50. March 2015. doi:10.1111/1462-2920.12509. PMID 24888970. https://pureadmin.qub.ac.uk/ws/files/18054125/Schmerk_EMI_2014_0306_revised_nofields.pdf.
- ↑ 33.0 33.1 "Identification of a methylase required for 2-methylhopanoid production and implications for the interpretation of sedimentary hopanes". Proceedings of the National Academy of Sciences of the United States of America 107 (19): 8537–42. May 2010. doi:10.1073/pnas.0912949107. PMID 20421508. Bibcode: 2010PNAS..107.8537W.
- ↑ 34.0 34.1 34.2 "Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production". Proceedings of the National Academy of Sciences of the United States of America 109 (32): 12905–10. August 2012. doi:10.1073/pnas.1208255109. PMID 22826256. Bibcode: 2012PNAS..10912905W.
- ↑ "A distinct pathway for tetrahymanol synthesis in bacteria". Proceedings of the National Academy of Sciences of the United States of America 112 (44): 13478–83. November 2015. doi:10.1073/pnas.1511482112. PMID 26483502. Bibcode: 2015PNAS..11213478B.
- ↑ "Hopanoids. 1. Geohopanoids: the most abundant natural products on Earth?". Accounts of Chemical Research 25 (9): 398–402. September 1992. doi:10.1021/ar00021a003.
- ↑ 37.0 37.1 Summons, Roger E.; Lincoln, Sara A. (2012-03-30). "Biomarkers: Informative Molecules for Studies in Geobiology". Fundamentals of Geobiology. John Wiley & Sons, Ltd. pp. 269–296. doi:10.1002/9781118280874.ch15. ISBN 978-1-118-28087-4.
- ↑ Knoll, Andrew H. (2003). Life on a young planet: the first three billion years of evolution on Earth. Princeton, N.J.: Princeton University Press. ISBN 0-691-00978-3. OCLC 50604948.
- ↑ "Biomarker evidence for green and purple sulphur bacteria in a stratified Palaeoproterozoic sea". Nature 437 (7060): 866–70. October 2005. doi:10.1038/nature04068. PMID 16208367. Bibcode: 2005Natur.437..866B.
- ↑ "Paleoproterozoic sterol biosynthesis and the rise of oxygen". Nature 543 (7645): 420–423. March 2017. doi:10.1038/nature21412. PMID 28264195. Bibcode: 2017Natur.543..420G.
- ↑ 41.0 41.1 Ourisson, Guy; Albrecht, Pierre; Rohmer, Michel (1982-07-01). "Predictive microbial biochemistry — from molecular fossils to procaryotic membranes" (in en). Trends in Biochemical Sciences 7 (7): 236–239. doi:10.1016/0968-0004(82)90028-7. ISSN 0968-0004.
- ↑ "Hypotheses for the origin and early evolution of triterpenoid cyclases". Geobiology 5 (1): 19–34. 2007. doi:10.1111/j.1472-4669.2007.00096.x. PMID 36298871.
- ↑ Summons, Roger E.; Jahnke, Linda L.; Hope, Janet M.; Logan, Graham A. (August 1999). "2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis" (in en). Nature 400 (6744): 554–557. doi:10.1038/23005. ISSN 1476-4687. PMID 10448856. https://www.nature.com/articles/23005.
- ↑ Brocks, J. J. (1999-08-13). "Archean Molecular Fossils and the Early Rise of Eukaryotes". Science 285 (5430): 1033–1036. doi:10.1126/science.285.5430.1033. PMID 10446042. https://www.science.org/doi/10.1126/science.285.5430.1033.
- ↑ French, Katherine L.; Hallmann, Christian; Hope, Janet M.; Schoon, Petra L.; Zumberge, J. Alex; Hoshino, Yosuke; Peters, Carl A.; George, Simon C. et al. (2015-05-12). "Reappraisal of hydrocarbon biomarkers in Archean rocks" (in en). Proceedings of the National Academy of Sciences 112 (19): 5915–5920. doi:10.1073/pnas.1419563112. ISSN 0027-8424. PMID 25918387.
- ↑ Kuypers, Marcel M.M.; van Breugel, Yvonne; Schouten, Stefan; Erba, Elisabetta; Sinninghe Damsté, Jaap S. (2004). "N2-fixing cyanobacteria supplied nutrient N for Cretaceous oceanic anoxic events" (in en). Geology 32 (10): 853. doi:10.1130/G20458.1. ISSN 0091-7613. https://pubs.geoscienceworld.org/geology/article/32/10/853-856/29362.
- ↑ Haddad, Emily E.; Tuite, Michael L.; Martinez, Aaron M.; Williford, Kenneth; Boyer, Diana L.; Droser, Mary L.; Love, Gordon D. (August 2016). "Lipid biomarker stratigraphic records through the Late Devonian Frasnian/Famennian boundary: Comparison of high- and low-latitude epicontinental marine settings" (in en). Organic Geochemistry 98: 38–53. doi:10.1016/j.orggeochem.2016.05.007.
- ↑ Marynowski, Leszek; Rakociński, Michał; Borcuch, Ewelina; Kremer, Barbara; Schubert, Brian A.; Jahren, A. Hope (June 2011). "Molecular and petrographic indicators of redox conditions and bacterial communities after the F/F mass extinction (Kowala, Holy Cross Mountains, Poland)" (in en). Palaeogeography, Palaeoclimatology, Palaeoecology 306 (1–2): 1–14. doi:10.1016/j.palaeo.2011.03.018. https://linkinghub.elsevier.com/retrieve/pii/S0031018211001428.
- ↑ "Biosynthesis of 2-methylbacteriohopanepolyols by an anoxygenic phototroph". Proceedings of the National Academy of Sciences of the United States of America 104 (38): 15099–104. September 2007. doi:10.1073/pnas.0704912104. PMID 17848515. Bibcode: 2007PNAS..10415099R.
- ↑ Ricci, J. N.; Michel, A. J.; Newman, D. K. (May 2015). "Phylogenetic analysis of HpnP reveals the origin of 2-methylhopanoid production in Alphaproteobacteria" (in en). Geobiology 13 (3): 267–277. doi:10.1111/gbi.12129. PMID 25630231. http://doi.wiley.com/10.1111/gbi.12129.
- ↑ "Cyanobacterial bacteriohopanepolyol signatures from cultures and natural environmental settings" (in en). Organic Geochemistry 39 (2): 232–263. 2008-02-01. doi:10.1016/j.orggeochem.2007.08.006.
- ↑ 52.0 52.1 Elling, Felix J.; Hemingway, Jordon D.; Evans, Thomas W.; Kharbush, Jenan J.; Spieck, Eva; Summons, Roger E.; Pearson, Ann (2020-12-29). "Vitamin B 12 -dependent biosynthesis ties amplified 2-methylhopanoid production during oceanic anoxic events to nitrification" (in en). Proceedings of the National Academy of Sciences 117 (52): 32996–33004. doi:10.1073/pnas.2012357117. ISSN 0027-8424. PMID 33318211.
- ↑ Ricci, Jessica N; Coleman, Maureen L; Welander, Paula V; Sessions, Alex L; Summons, Roger E; Spear, John R; Newman, Dianne K (March 2014). "Diverse capacity for 2-methylhopanoid production correlates with a specific ecological niche" (in en). The ISME Journal 8 (3): 675–684. doi:10.1038/ismej.2013.191. ISSN 1751-7362. PMID 24152713.
- ↑ 54.0 54.1 Zundel, Magali; Rohmer, Michel (July 1985). "Prokaryotic triterpenoids. 3. The biosynthesis of 2beta-methylhopanoids and 3beta-methylhopanoids of Methylobacterium organophilum and Acetobacter pasteurianus ssp. pasteurianus" (in en). European Journal of Biochemistry 150 (1): 35–39. doi:10.1111/j.1432-1033.1985.tb08984.x. ISSN 0014-2956. PMID 3926496.
- ↑ Elvert, Marcus; Niemann, Helge (February 2008). "Occurrence of unusual steroids and hopanoids derived from aerobic methanotrophs at an active marine mud volcano" (in en). Organic Geochemistry 39 (2): 167–177. doi:10.1016/j.orggeochem.2007.11.006. https://linkinghub.elsevier.com/retrieve/pii/S0146638007002604.
- ↑ "Squalene hopene cyclases are protonases for stereoselective Brønsted acid catalysis". Nature Chemical Biology 11 (2): 121–6. February 2015. doi:10.1038/nchembio.1719. PMID 25503928.
- ↑ Newman DK, Kulkarni G, Belin BJ, "Hopanoids producing bacteria and related biofertilizers, compositions, methods and systems", US patent 2017107160, issued 2016-10-19, assigned to California Institute of Technology
 |
 KSF
KSF