Marine prokaryotes
Topic: Biology
 From HandWiki - Reading time: 45 min
From HandWiki - Reading time: 45 min
| Part of a series of overviews on |
| Marine life |
|---|
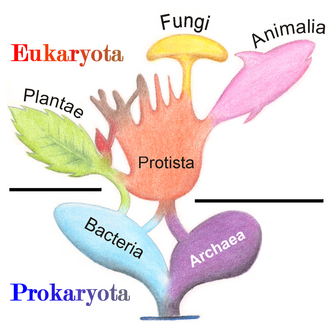
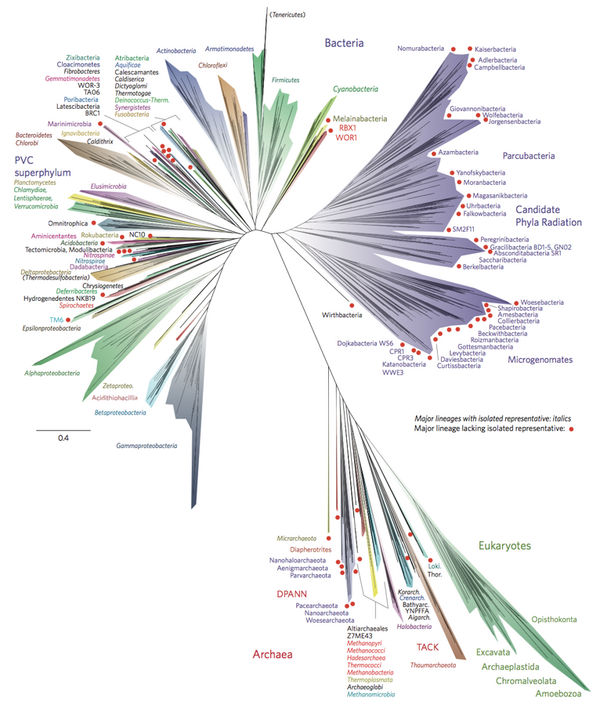
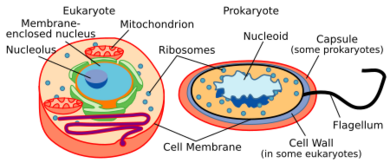
Marine prokaryotes are marine bacteria and marine archaea. They are defined by their habitat as prokaryotes that live in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. All cellular life forms can be divided into prokaryotes and eukaryotes. Eukaryotes are organisms whose cells have a nucleus enclosed within membranes, whereas prokaryotes are the organisms that do not have a nucleus enclosed within a membrane.[1][2][3] The three-domain system of classifying life adds another division: the prokaryotes are divided into two domains of life, the microscopic bacteria and the microscopic archaea, while everything else, the eukaryotes, become the third domain.[4]
Prokaryotes play important roles in ecosystems as decomposers recycling nutrients. Some prokaryotes are pathogenic, causing disease and even death in plants and animals.[5] Marine prokaryotes are responsible for significant levels of the photosynthesis that occurs in the ocean, as well as significant cycling of carbon and other nutrients.[6]
Prokaryotes live throughout the biosphere. In 2018 it was estimated the total biomass of all prokaryotes on the planet was equivalent to 77 billion tonnes of carbon (77 Gt C). This is made up of 7 Gt C for archaea and 70 Gt C for bacteria. These figures can be contrasted with the estimate for the total biomass for animals on the planet, which is about 2 Gt C, and the total biomass of humans, which is 0.06 Gt C.[7] This means archaea collectively have over 100 times the collective biomass of humans, and bacteria over 1000 times.
There is no clear evidence of life on Earth during the first 600 million years of its existence. When life did arrive, it was dominated for 3,200 million years by the marine prokaryotes. More complex life, in the form of crown eukaryotes, didn't appear until the Cambrian explosion a mere 500 million years ago.[8]
Evolution
The Earth is about 4.54 billion years old.[9][10][11] The earliest undisputed evidence of life on Earth dates from at least 3.5 billion years ago,[12][13] during the Eoarchean Era after a geological crust started to solidify following the earlier molten Hadean Eon. Microbial mat fossils have been found in 3.48 billion-year-old sandstone in Western Australia.[14][15]
Past species have also left records of their evolutionary history. Fossils, along with the comparative anatomy of present-day organisms, constitute the morphological, or anatomical, record.[16] By comparing the anatomies of both modern and extinct species, paleontologists can infer the lineages of those species. However, this approach is most successful for organisms that had hard body parts, such as shells, bones or teeth. Further, as prokaryotes such as bacteria and archaea share a limited set of common morphologies, their fossils do not provide information on their ancestry.
Prokaryotes inhabited the Earth from approximately 3–4 billion years ago.[17][18] No obvious changes in morphology or cellular organisation occurred in these organisms over the next few billion years.[19] The eukaryotic cells emerged between 1.6 and 2.7 billion years ago. The next major change in cell structure came when bacteria were engulfed by eukaryotic cells, in a cooperative association called endosymbiosis.[20][21] The engulfed bacteria and the host cell then underwent coevolution, with the bacteria evolving into either mitochondria or hydrogenosomes.[22] Another engulfment of cyanobacterial-like organisms led to the formation of chloroplasts in algae and plants.[23]
The history of life was that of the unicellular prokaryotes and eukaryotes until about 610 million years ago when multicellular organisms began to appear in the oceans in the Ediacaran period.[17][26] The evolution of multicellularity occurred in multiple independent events, in organisms as diverse as sponges, brown algae, cyanobacteria, slime moulds and myxobacteria.[27] In 2016 scientists reported that, about 800 million years ago, a minor genetic change in a single molecule called GK-PID may have allowed organisms to go from a single cell organism to one of many cells.[28]
Soon after the emergence of these first multicellular organisms, a remarkable amount of biological diversity appeared over a span of about 10 million years, in an event called the Cambrian explosion. Here, the majority of types of modern animals appeared in the fossil record, as well as unique lineages that subsequently became extinct.[29] Various triggers for the Cambrian explosion have been proposed, including the accumulation of oxygen in the atmosphere from photosynthesis.[30]
Background

The words prokaryote and eukaryote come from the Greek where pro means "before", eu means "well" or "true", and karyon means "nut", "kernel" or "nucleus".[31][32][33] So etymologically, prokaryote means "before nucleus" and eukaryote means "true nucleus".
The division of life forms between prokaryotes and eukaryotes was firmly established by the microbiologists Roger Stanier and C. B. van Niel in their 1962 paper, The concept of a bacterium.[34] One reason for this classification was so what was then often called blue-green algae (now called cyanobacteria) would cease to be classified as plants but grouped with bacteria.
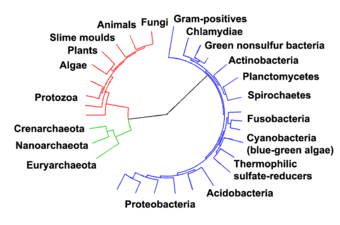
In 1990 Carl Woese et al. introduced the three-domain system.[35][36] The prokaryotes were split into two domains, the archaea and the bacteria, while the eukaryotes become a domain in their own right. The key difference from earlier classifications is the splitting of archaea from bacteria.

The earliest evidence for life on earth comes from biogenic carbon signatures and stromatolite fossils discovered in 3.7 billion-year-old rocks.[37][38] In 2015, possible "remains of biotic life" were found in 4.1 billion-year-old rocks.[39][40] In 2017 putative evidence of possibly the oldest forms of life on Earth was reported in the form of fossilized microorganisms discovered in hydrothermal vent precipitates that may have lived as early as 4.28 billion years ago, not long after the oceans formed 4.4 billion years ago, and not long after the formation of the Earth 4.54 billion years ago.[41][42]
Microbial mats of coexisting bacteria and archaea were the dominant form of life in the early Archean Eon and many of the major steps in early evolution are thought to have taken place in this environment.[43] The evolution of photosynthesis around 3.5 Ga resulted in a buildup of its waste product oxygen in the atmosphere, leading to the great oxygenation event beginning around 2.4 Ga.[44]
The earliest evidence of eukaryotes dates from 1.85 Ga,[45][46] and while they may have been present earlier, their diversification accelerated when they started using oxygen in their metabolism. Later, around 1.7 Ga, multicellular organisms began to appear, with differentiated cells performing specialised functions.[47]
A stream of airborne microorganisms, including prokaryotes, circles the planet above weather systems but below commercial air lanes.[51] Some peripatetic microorganisms are swept up from terrestrial dust storms, but most originate from marine microorganisms in sea spray. In 2018, scientists reported that hundreds of millions of viruses and tens of millions of bacteria are deposited daily on every square meter around the planet.[52][53]
Microscopic life undersea is diverse and still poorly understood, such as for the role of viruses in marine ecosystems.[54] Most marine viruses are bacteriophages, which are harmless to plants and animals, but are essential to the regulation of saltwater and freshwater ecosystems.[55] They infect and destroy bacteria and archaea in aquatic microbial communities, and are the most important mechanism of recycling carbon in the marine environment. The organic molecules released from the dead bacterial cells stimulate fresh bacterial and algal growth.[56] Viral activity may also contribute to the biological pump, the process whereby carbon is sequestered in the deep ocean.[57]
Marine bacteria
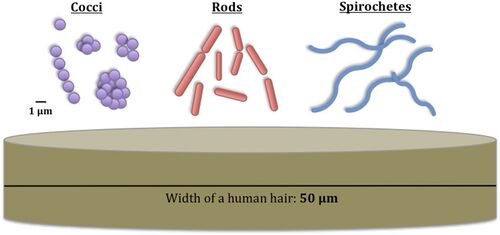
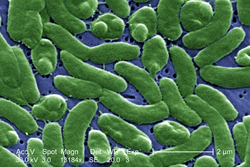
Bacteria constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a number of shapes, ranging from spheres to rods and spirals. Bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste,[61] and the deep portions of Earth's crust. Bacteria also live in symbiotic and parasitic relationships with plants and animals.
Once regarded as plants constituting the class Schizomycetes, bacteria are now classified as prokaryotes. Unlike cells of animals and other eukaryotes, bacterial cells do not contain a nucleus and rarely harbour membrane-bound organelles. Although the term bacteria traditionally included all prokaryotes, the scientific classification changed after the discovery in the 1990s that prokaryotes consist of two very different groups of organisms that evolved from an ancient common ancestor. These evolutionary domains are called Bacteria and Archaea.[62]
The ancestors of modern bacteria were unicellular microorganisms that were the first forms of life to appear on Earth, about 4 billion years ago. For about 3 billion years, most organisms were microscopic, and bacteria and archaea were the dominant forms of life.[63][64] Although bacterial fossils exist, such as stromatolites, their lack of distinctive morphology prevents them from being used to examine the history of bacterial evolution, or to date the time of origin of a particular bacterial species. However, gene sequences can be used to reconstruct the bacterial phylogeny, and these studies indicate that bacteria diverged first from the archaeal/eukaryotic lineage.[65] Bacteria were also involved in the second great evolutionary divergence, that of the archaea and eukaryotes. Here, eukaryotes resulted from the entering of ancient bacteria into endosymbiotic associations with the ancestors of eukaryotic cells, which were themselves possibly related to the Archaea.[21][66] This involved the engulfment by proto-eukaryotic cells of alphaproteobacterial symbionts to form either mitochondria or hydrogenosomes, which are still found in all known Eukarya. Later on, some eukaryotes that already contained mitochondria also engulfed cyanobacterial-like organisms. This led to the formation of chloroplasts in algae and plants. There are also some algae that originated from even later endosymbiotic events. Here, eukaryotes engulfed a eukaryotic algae that developed into a "second-generation" plastid.[67][68] This is known as secondary endosymbiosis.
Bacteria grow to a fixed size and then reproduce through binary fission, a form of asexual reproduction.[69] Under optimal conditions, bacteria can grow and divide extremely rapidly, and bacterial populations can double as quickly as every 9.8 minutes.[70]
Pelagibacter ubique and its relatives may be the most abundant microorganisms in the ocean, and it has been claimed that they are possibly the most abundant bacteria in the world. They make up about 25% of all microbial plankton cells, and in the summer they may account for approximately half the cells present in temperate ocean surface water. The total abundance of P. ubique and relatives is estimated to be about 2 × 1028 microbes.[71] However, it was reported in Nature in February 2013 that the bacteriophage HTVC010P, which attacks P. ubique, has been discovered and is probably the most common organism on the planet.[72][73]
Roseobacter is also one of the most abundant and versatile microorganisms in the ocean. They are diversified across different types of marine habitats, from coastal to open oceans and from sea ice to sea floor, and make up about 25% of coastal marine bacteria. Members of the Roseobacter genus play important roles in marine biogeochemical cycles and climate change, processing a significant portion of the total carbon in the marine environment. They form symbiotic relationships which allow them to degrade aromatic compounds and uptake trace metals. They are widely used in aquaculture and quorum sensing. During algal blooms, 20-30% of the prokaryotic community are Roseobacter.[74][75]
The largest known bacterium, the marine Thiomargarita namibiensis, can be visible to the naked eye and sometimes attains 0.75 mm (750 μm).[76][77]
Cyanobacteria
Cyanobacteria were the first organisms to evolve an ability to turn sunlight into chemical energy. They form a phylum (division) of bacteria which range from unicellular to filamentous and include colonial species. They are found almost everywhere on earth: in damp soil, in both freshwater and marine environments, and even on Antarctic rocks.[79] In particular, some species occur as drifting cells floating in the ocean, and as such were amongst the first of the phytoplankton.
The first primary producers that used photosynthesis were oceanic cyanobacteria about 2.3 billion years ago.[80][81] The release of molecular oxygen by cyanobacteria as a by-product of photosynthesis induced global changes in the Earth's environment. Because oxygen was toxic to most life on Earth at the time, this led to the near-extinction of oxygen-intolerant organisms, a dramatic change which redirected the evolution of the major animal and plant species.[82]

interact with bacteria to acquire iron from dust
b. Trichodesmium can establish massive blooms in nutrient poor ocean regions with high dust deposition, partly due to their unique ability to capture dust, center it, and subsequently dissolve it.
c. Proposed dust-bound Fe acquisition pathway: Bacteria residing within the colonies produce siderophores (c-I) that react with the dust particles in the colony core and generate dissolved Fe (c-II). This dissolved Fe, complexed by siderophores, is then acquired by both Trichodesmium and its resident bacteria (c-III), resulting in a mutual benefit to both partners of the consortium.[83]
-
Bloom of the filamentous cyanobacteria Trichodesmium
-
Synechococcus, a widespread marine cyanobacterium
-
Carboxysomes appearing as polyhedral dark structures within a species of Synechococcus
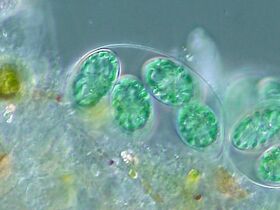
The tiny (0.6 µm) marine cyanobacterium Prochlorococcus, discovered in 1986, forms today an important part of the base of the ocean food chain and accounts for much of the photosynthesis of the open ocean[85] and an estimated 20% of the oxygen in the Earth's atmosphere.[86] It is possibly the most plentiful genus on Earth: a single millilitre of surface seawater may contain 100,000 cells or more.[87]
Originally, biologists classified cyanobacteria as an algae, and referred to it as "blue-green algae". The more recent view is that cyanobacteria are bacteria, and hence are not even in the same Kingdom as algae. Most authorities exclude all prokaryotes, and hence cyanobacteria from the definition of algae.[88][89]
Other bacteria
Other marine bacteria, apart from cyanobacteria, are ubiquitous or can play important roles in the ocean. These include the opportunistic copiotroph, Alteromonas macleodii.[90][91]
Marine archaea

The archaea (Greek for ancient[93]) constitute a domain and kingdom of single-celled microorganisms. These microbes are prokaryotes, meaning they have no cell nucleus or any other membrane-bound organelles in their cells.
Archaea were initially classified as bacteria, but this classification is outdated.[94] Archaeal cells have unique properties separating them from the other two domains of life, Bacteria and Eukaryota. The Archaea are further divided into multiple recognized phyla. Classification is difficult because the majority have not been isolated in the laboratory and have only been detected by analysis of their nucleic acids in samples from their environment.
Bacteria and archaea are generally similar in size and shape, although a few archaea have very strange shapes, such as the flat and square-shaped cells of Haloquadratum walsbyi.[95] Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably the enzymes involved in transcription and translation. Other aspects of archaeal biochemistry are unique, such as their reliance on ether lipids in their cell membranes, such as archaeols. Archaea use more energy sources than eukaryotes: these range from organic compounds, such as sugars, to ammonia, metal ions or even hydrogen gas. Salt-tolerant archaea (the Haloarchaea) use sunlight as an energy source, and other species of archaea fix carbon; however, unlike plants and cyanobacteria, no known species of archaea does both. Archaea reproduce asexually by binary fission, fragmentation, or budding; unlike bacteria and eukaryotes, no known species forms spores.
Archaea are particularly numerous in the oceans, and the archaea in plankton may be one of the most abundant groups of organisms on the planet. Archaea are a major part of Earth's life and may play roles in both the carbon cycle and the nitrogen cycle. Thermoproteota (also called Crenarchaeota or eocytes) are a phylum of archaea thought to be very abundant in marine environments and one of the main contributors to the fixation of carbon.[96]
-
Eocytes may be the most abundant of marine archaea
-
Halobacteria, found in water near saturated with salt, are now recognised as archaea.
-
Flat, square-shaped cells of the archaea Haloquadratum walsbyi
-
Methanosarcina barkeri, a marine archaea that produces methane
-
Thermophiles, such as Pyrolobus fumarii, survive well over 100 °C
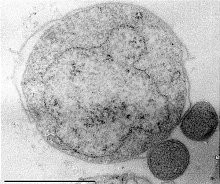
Nanoarchaeum equitans is a species of marine archaea discovered in 2002 in a hydrothermal vent. It is a thermophile that grows in temperatures at about 80 degrees Celsius. Nanoarchaeum appears to be an obligate symbiont on the archaeon Ignicoccus. It must stay in contact with the host organism to survive since Nanoarchaeum equitans cannot synthesize lipids but obtains them from its host. Its cells are only 400 nm in diameter, making it one of the smallest known cellular organisms, and the smallest known archaeon.[97][98]
Marine archaea have been classified as follows:[99][100][101][102][103]
- Marine Group I (MG-I or MGI): marine Nitrososphaerota with subgroups Ia (aka I.a) up to Id
- Marine Group II (MG-II): marine Euryarchaeota, order Poseidoniales[104] with subgroups IIa up to IId (IIa resembling Poseidoniaceae, IIb resembling Thalassarchaceae)
Viruses parasiting MGII are classified as magroviruses - Marine Group III (MG-III): also marine Euryarchaeota, Marine Benthic Group D[105]
- Marine Group IV (MG-IV): also marine Euryarchaeota[106]
Trophic mode
Prokaryote metabolism is classified into nutritional groups on the basis of three major criteria: the source of energy, the electron donors used, and the source of carbon used for growth.[107][108]
| Nutritional type | Source of energy | Source of carbon | Examples |
|---|---|---|---|
| Phototrophs | Sunlight | Organic compounds (photoheterotrophs) or carbon fixation (photoautotrophs) | "Cyanobacteria", Green sulfur bacteria, Chloroflexota, or Purple bacteria |
| Lithotrophs | Inorganic compounds | Organic compounds (lithoheterotrophs) or carbon fixation (lithoautotrophs) | Thermodesulfobacteriota, Hydrogenophilaceae, or Nitrospirota |
| Organotrophs | Organic compounds | Organic compounds (chemoheterotrophs) or carbon fixation (chemoautotrophs) | Bacillus, Clostridium or Enterobacteriaceae |
Marine prokaryotes have diversified greatly throughout their long existence. The metabolism of prokaryotes is far more varied than that of eukaryotes, leading to many highly distinct prokaryotic types. For example, in addition to using photosynthesis or organic compounds for energy, as eukaryotes do, marine prokaryotes may obtain energy from inorganic compounds such as hydrogen sulfide. This enables marine prokaryotes to thrive as extremophiles in harsh environments as cold as the ice surface of Antarctica, studied in cryobiology, as hot as undersea hydrothermal vents, or in high saline conditions as (halophiles).[109] Some marine prokaryotes live symbiotically in or on the bodies of other marine organisms.

- Phototrophy is a particularly significant marker that should always play a primary role in bacterial classification.[110]
- Aerobic anoxygenic phototrophic bacteria (AAPBs) are widely distributed marine plankton that may constitute over 10% of the open ocean microbial community. Marine AAPBs are classified in two marine (Erythrobacter and Roseobacter) genera. They can be particularly abundant in oligotrophic conditions where they were found to be 24% of the community.[111] These are heterotrophic organisms that use light to produce energy, but are unable to utilise carbon dioxide as their primary carbon source. Most are obligately aerobic, meaning they require oxygen to grow. Current data suggests that marine bacteria have generation times of several days, whereas new evidence exists that shows AAPB to have a much shorter generation time.[112] Coastal/shelf waters often have greater amounts of AAPBs, some as high as 13.51% AAPB%. Phytoplankton also affect AAPB%, but little research has been performed in this area.[113] They can also be abundant in various oligotrophic conditions, including the most oligotrophic regime of the world ocean.[114] They are globally distributed in the euphotic zone and represent a hitherto unrecognized component of the marine microbial community that appears to be critical to the cycling of both organic and inorganic carbon in the ocean.[115]
- Purple bacteria:
- Zetaproteobacteria: are iron-oxidizing neutrophilic chemolithoautotrophs, distributed worldwide in estuaries and marine habitats.
- Hydrogen oxidizing bacteria are facultative autotrophs that can be divided into aerobes and anaerobes. The former use hydrogen as an electron donor and oxygen as an acceptor while the latter use sulphate or nitrogen dioxide as electron acceptors.[116]
Motility
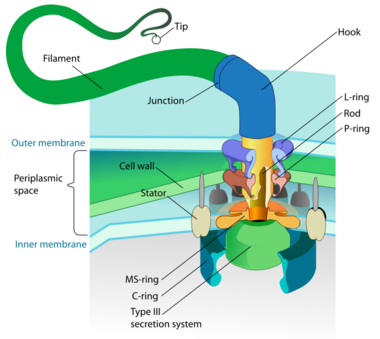
Motility is the ability of an organism to move independently, using metabolic energy.
Flagellar motility
Prokaryotes, both bacteria and archaea, primarily use flagella for locomotion.
- Bacterial flagella are helical filaments, each with a rotary motor at its base which can turn clockwise or counterclockwise.[118][119][120] They provide two of several kinds of bacterial motility.[121][122]
- Archaeal flagella are called archaella, and function in much the same way as bacterial flagella. Structurally the archaellum is superficially similar to a bacterial flagellum, but it differs in many details and is considered non-homologous.[123][117]
The rotary motor model used by bacteria uses the protons of an electrochemical gradient in order to move their flagella. Torque in the flagella of bacteria is created by particles that conduct protons around the base of the flagellum. The direction of rotation of the flagella in bacteria comes from the occupancy of the proton channels along the perimeter of the flagellar motor.[124]
Some eukaryotic cells also use flagella — and they can be found in some protists and plants as well as animal cells. Eukaryotic flagella are complex cellular projections that lash back and forth, rather than in a circular motion. Prokaryotic flagella use a rotary motor, and the eukaryotic flagella use a complex sliding filament system. Eukaryotic flagella are ATP-driven, while prokaryotic flagella can be ATP-driven (archaea) or proton-driven (bacteria).[125]
Twitching motility
Twitching motility is a form of crawling bacterial motility used to move over surfaces. Twitching is mediated by the activity of hair-like filaments called type IV pili which extend from the cell's exterior, bind to surrounding solid substrates and retract, pulling the cell forwards in a manner similar to the action of a grappling hook.[126][127][128] The name twitching motility is derived from the characteristic jerky and irregular motions of individual cells when viewed under the microscope.[129]
Gliding motility
Gliding motility is a type of translocation that is independent of propulsive structures such as flagella or pili.[130] Gliding allows microorganisms to travel along the surface of low aqueous films. The mechanisms of this motility are only partially known. The speed of gliding varies between organisms, and the reversal of direction is seemingly regulated by some sort of internal clock.[131] For example, the apicomplexans are able to travel at fast rates between 1–10 μm/s. In contrast Myxococcus xanthus bacteria glide at a rate of 5 μm/min.[132][133]
Swarming motility
Swarming motility is a rapid (2–10 μm/s) and coordinated translocation of a bacterial population across solid or semi-solid surfaces,[134] and is an example of bacterial multicellularity and swarm behaviour. Swarming motility was first reported in 1972 by Jorgen Henrichsen.[135]
Non-motile
Non-motile species lack the ability and structures that would allow them to propel themselves, under their own power, through their environment. When non-motile bacteria are cultured in a stab tube, they only grow along the stab line. If the bacteria are mobile, the line will appear diffuse and extend into the medium.[136]
Taxis: Directed motion
Magnetotaxis
Magnetotactic bacteria orient themselves along the magnetic field lines of Earth's magnetic field.[138] This alignment is believed to aid these organisms in reaching regions of optimal oxygen concentration.[139] To perform this task, these bacteria have biomineralised organelles called magnetosomes that contain magnetic crystals. The biological phenomenon of microorganisms tending to move in response to the environment's magnetic characteristics is known as magnetotaxis. However, this term is misleading in that every other application of the term taxis involves a stimulus-response mechanism. In contrast to the magnetoreception of animals, the bacteria contain fixed magnets that force the bacteria into alignment—even dead cells are dragged into alignment, just like a compass needle.[139]
Marine environments are generally characterized by low concentrations of nutrients kept in steady or intermittent motion by currents and turbulence. Marine bacteria have developed strategies, such as swimming and using directional sensing–response systems, to migrate towards favorable places in the nutrient gradients. Magnetotactic bacteria utilize Earth's magnetic field to facilitate downward swimming into the oxic–anoxic interface, which is the most favorable place for their persistence and proliferation, in chemically stratified sediments or water columns.[140]
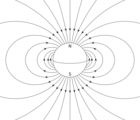
Depending on their latitude and whether the bacteria are north or south of the equator, the Earth's magnetic field has one of the two possible polarities, and a direction that points with varying angles into the ocean depths, and away from the generally more oxygen rich surface. Aerotaxis is the response by which bacteria migrate to an optimal oxygen concentration in an oxygen gradient. Various experiments have clearly shown that magnetotaxis and aerotaxis work in conjunction in magnetotactic bacteria. It has been shown that, in water droplets, one-way swimming magnetotactic bacteria can reverse their swimming direction and swim backwards under reducing conditions (less than optimal oxygen concentration), as opposed to oxic conditions (greater than optimal oxygen concentration).
Regardless of their morphology, all magnetotactic bacteria studied so far are motile by means of flagella.[141] Marine magnetotactic bacteria in particular tend to possess an elaborate flagellar apparatus which can involve up to tens of thousands of flagella. However, despite extensive research in recent years, it has yet to be established whether magnetotactic bacteria steer their flagellar motors in response to their alignment in magnetic fields.[140] Symbiosis with magnetotactic bacteria has been proposed as the explanation for magnetoreception in some marine protists.[142] Research is underway on whether a similar relationship may underlie magnetoreception in vertebrates as well.[143] The oldest unambiguous magnetofossils come from the Cretaceous chalk beds of southern England,[144] though less certain reports of magnetofossils extend to 1.9 billion years old Gunflint Chert.[145]
Gas vacuoles
Some marine prokaryotes possess gas vacuoles. Gas vacuole are nanocompartments freely permeable to gas which allow marine bacteria and archaea to control their buoyancy. They take the form of spindle-shaped membrane-bound vesicles, and are found in some plankton prokaryotes, including some Cyanobacteria.[146] Positive buoyancy is needed to keep the cells in the upper reaches of the water column, so that they can continue to perform photosynthesis. Gas vacuoles are made up of a shell of protein that has a highly hydrophobic inner surface, making it impermeable to water (and stopping water vapour from condensing inside) but permeable to most gases. Because the gas vesicle is a hollow cylinder, it is liable to collapse when the surrounding pressure increases. Natural selection has fine tuned the structure of the gas vesicle to maximise its resistance to buckling, including an external strengthening protein, GvpC, rather like the green thread in a braided hosepipe. There is a simple relationship between the diameter of the gas vesicle and pressure at which it will collapse – the wider the gas vesicle the weaker it becomes. However, wider gas vesicles are more efficient, providing more buoyancy per unit of protein than narrow gas vesicles. Different species produce gas vesicle of different diameter, allowing them to colonise different depths of the water column (fast growing, highly competitive species with wide gas vesicles in the top most layers; slow growing, dark-adapted, species with strong narrow gas vesicles in the deeper layers).
The cell achieves its height in the water column by synthesising gas vesicles. As the cell rises up, it is able to increase its carbohydrate load through increased photosynthesis. Too high and the cell will suffer photobleaching and possible death, however, the carbohydrate produced during photosynthesis increases the cell's density, causing it to sink. The daily cycle of carbohydrate build-up from photosynthesis and carbohydrate catabolism during dark hours is enough to fine-tune the cell's position in the water column, bring it up toward the surface when its carbohydrate levels are low and it needs to photosynthesis, and allowing it to sink away from the harmful UV radiation when the cell's carbohydrate levels have been replenished. An extreme excess of carbohydrate causes a significant change in the internal pressure of the cell, which causes the gas vesicles to buckle and collapse and the cell to sink out.
Large vacuoles are found in three genera of filamentous sulfur bacteria, the Thioploca, Beggiatoa and Thiomargarita. The cytosol is extremely reduced in these genera and the vacuole can occupy between 40 and 98% of the cell.[147] The vacuole contains high concentrations of nitrate ions and is therefore thought to be a storage organelle.[148]
Bioluminescence
Bioluminescent bacteria are light-producing bacteria that are predominantly present in sea water, marine sediments, the surface of decomposing fish and in the gut of marine animals. While not as common, bacterial bioluminescence is also found in terrestrial and freshwater bacteria.[126] These bacteria may be free living (such as Vibrio harveyi) or in symbiosis with animals such as the Hawaiian Bobtail squid (Aliivibrio fischeri) or terrestrial nematodes (Photorhabdus luminescens). The host organisms provide these bacteria a safe home and sufficient nutrition. In exchange, the hosts use the light produced by the bacteria for camouflage, prey and/or mate attraction. Bioluminescent bacteria have evolved symbiotic relationships with other organisms in which both participants benefit close to equally.[150] Another possible reason bacteria use luminescence reaction is for quorum sensing, an ability to regulate gene expression in response to bacterial cell density.[151]
The Hawaiian bobtail squid lives in symbiosis with the bioluminescent bacteria Aliivibrio fischeri which inhabits a special light organ in the squid's mantle. The bacteria are fed sugar and amino acid by the squid and in return hide the squid's silhouette when viewed from below, counter-illuminating it by matching the amount of light hitting the top of the mantle.[152] The squid serves as a model organism for animal-bacterial symbiosis and its relationship with the bacteria has been widely studied.
Vibrio harveyi is a rod-shaped, motile (via polar flagella) bioluminescent bacterium which grows optimally between 30° and 35 °C. It can be found free-swimming in tropical marine waters, commensally in the gut microflora of marine animals, and as both a primary and opportunistic pathogen of a number of marine animals.[153] It is thought to be the cause of the milky seas effect, where a uniform blue glow is emitted from seawater during the night. Some glows can cover nearly 6,000 sq mi (16,000 km2).
Microbial rhodopsin
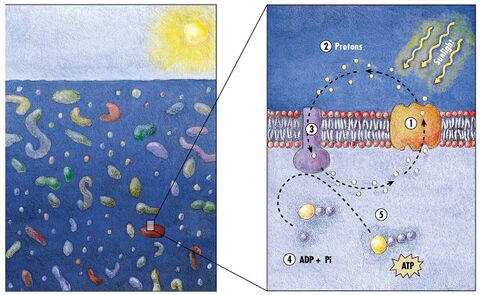
(2) it changes its configuration so a proton is expelled from the cell
(3) the chemical potential causes the proton to flow back to the cell
(4) thus generating energy
(5) in the form of adenosine triphosphate.[154]
Phototrophic metabolism relies on one of three energy-converting pigments: chlorophyll, bacteriochlorophyll, and retinal. Retinal is the chromophore found in rhodopsins. The significance of chlorophyll in converting light energy has been written about for decades, but phototrophy based on retinal pigments is just beginning to be studied.[155]

In 2000 a team of microbiologists led by Edward DeLong made a crucial discovery in the understanding of the marine carbon and energy cycles. They discovered a gene in several species of bacteria[157][158] responsible for production of the protein rhodopsin, previously unheard of in bacteria. These proteins found in the cell membranes are capable of converting light energy to biochemical energy due to a change in configuration of the rhodopsin molecule as sunlight strikes it, causing the pumping of a proton from inside out and a subsequent inflow that generates the energy.[159] The archaeal-like rhodopsins have subsequently been found among different taxa, protists as well as in bacteria and archaea, though they are rare in complex multicellular organisms.[160][161][162]
Research in 2019 shows these "sun-snatching bacteria" are more widespread than previously thought and could change how oceans are affected by global warming. "The findings break from the traditional interpretation of marine ecology found in textbooks, which states that nearly all sunlight in the ocean is captured by chlorophyll in algae. Instead, rhodopsin-equipped bacteria function like hybrid cars, powered by organic matter when available — as most bacteria are — and by sunlight when nutrients are scarce."[163][155]
There is an astrobiological conjecture called the Purple Earth hypothesis which surmises that original life forms on Earth were retinal-based rather than chlorophyll-based, which would have made the Earth appear purple instead of green.[164][165]
Symbiosis
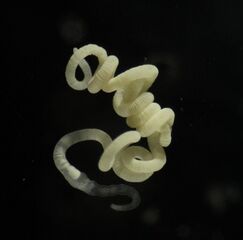
Some marine organisms have a symbiosis with bacteria or archaea. Pompeii worms live at great depths by hydrothermal vents at temperatures up to 80 °C. They have what appear to be hairy backs, but these "hairs" are actually colonies of bacteria such as Nautilia profundicola, which are thought to afford the worm some degree of insulation. Glands on the worm's back secrete a mucus on which the bacteria feed, a form of symbiosis.
-
The "hairy" backs of Pompeii worms are colonies of symbiotic bacteria
-
Hesiocaeca methanicola lives at great depths on methane ice and appear to survive in symbiosis with bacteria which metabolize the clathrate.[166]
-
Olavius algarvensis depends on five different species of symbiotic bacteria for its nutrition
-
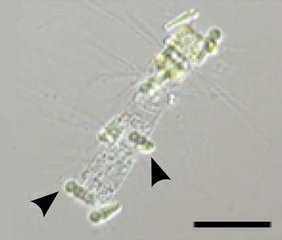
Epiphytic Calothrix cyanobacteria (arrows) in symbiosis with a Chaetoceros diatom. Scale bar 50 μm.
Endosymbiont bacteria are bacteria that live within the body or cells of another organism. Some types of cyanobacteria are endosymbiont and cyanobacteria have been found to possess genes that enable them to undergo nitrogen fixation.[168]
Organisms typically establish a symbiotic relationship due to their limited availability of resources in their habitat or due to a limitation of their food source. Symbiotic, chemosynthetic bacteria that have been discovered associated with mussels (Bathymodiolus) located near hydrothermal vents have a gene that enables them to utilize hydrogen as a source of energy, in preference to sulphur or methane as their energy source for production of energy.[169]
Olavius algarvensis is a worm which lives in coastal sediments in the Mediterranean and depends on symbiotic bacteria for its nutrition. It lives with five different species of bacteria located under its cuticle: two sulfide-oxidizing , two sulfate-reducing and one spirochaete. The symbiotic bacteria also allow the worm to use hydrogen and carbon monoxide as energy sources, and to metabolise organic compounds like malate and acetate.[170][171]
Astrangia poculata, the northern star coral, is a temperate stony coral, widely documented along the eastern coast of the United States. The coral can live with and without zooxanthellae (algal symbionts), making it an ideal model organism to study microbial community interactions associated with symbiotic state. However, the ability to develop primers and probes to more specifically target key microbial groups has been hindered by the lack of full length 16S rRNA sequences, since sequences produced by the Illumina platform are of insufficient length (approximately 250 base pairs) for the design of primers and probes.[172] In 2019, Goldsmith et al. demonstrated Sanger sequencing was capable of reproducing the biologically-relevant diversity detected by deeper next-generation sequencing, while also producing longer sequences useful to the research community for probe and primer design (see diagram on right).[173]
Roles in marine food webs
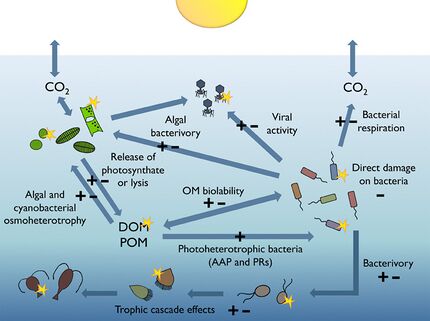
Most of the volume of the world ocean is in darkness. The processes occurring within the thin illuminated surface layer (the photic layer from the surface down to between 50 and 170 metres) are of major significance to the global biosphere. For example, the visible region of the solar spectrum (the so-called photosynthetically available radiation or PAR) reaching this sunlit layer fuels about half of the primary productivity of the planet, and is responsible for about half of the atmospheric oxygen necessary for most life on Earth.[175][176]
Heterotrophic bacterioplankton are main consumers of dissolved organic matter (DOM) in pelagic marine food webs, including the sunlit upper layers of the ocean. Their sensitivity to ultraviolet radiation (UVR), together with some recently discovered mechanisms bacteria have evolved to benefit from photosynthetically available radiation (PAR), suggest that natural sunlight plays a relevant, yet difficult to predict role in modulating bacterial biogeochemical functions in the oceans.[174]
Ocean surface habitats sit at the interface between the atmosphere and the ocean. The biofilm-like habitat at the surface of the ocean harbours surface-dwelling microorganisms, commonly referred to as neuston. This vast air–water interface sits at the intersection of major air–water exchange processes spanning more than 70% of the global surface area . Bacteria in the surface microlayer of the ocean, called bacterioneuston, are of interest due to practical applications such as air-sea gas exchange of greenhouse gases, production of climate-active marine aerosols, and remote sensing of the ocean.[177] Of specific interest is the production and degradation of surfactants (surface active materials) via microbial biochemical processes. Major sources of surfactants in the open ocean include phytoplankton,[178] terrestrial runoff, and deposition from the atmosphere.[177]

Unlike coloured algal blooms, surfactant-associated bacteria may not be visible in ocean colour imagery. Having the ability to detect these "invisible" surfactant-associated bacteria using synthetic aperture radar has immense benefits in all-weather conditions, regardless of cloud, fog, or daylight.[177] This is particularly important in very high winds, because these are the conditions when the most intense air-sea gas exchanges and marine aerosol production take place. Therefore, in addition to colour satellite imagery, SAR satellite imagery may provide additional insights into a global picture of biophysical processes at the boundary between the ocean and atmosphere, air-sea greenhouse gas exchanges and production of climate-active marine aerosols.[177]
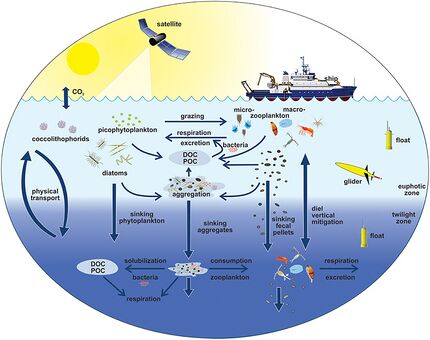
The diagram on the right shows links among the ocean's biological pump and the pelagic food web and the ability to sample these components remotely from ships, satellites, and autonomous vehicles. Light blue waters are the euphotic zone, while the darker blue waters represent the twilight zone.[179]
Roles in biogeochemical cycling
Archaea recycle elements such as carbon, nitrogen, and sulfur through their various habitats.[180] Archaea carry out many steps in the nitrogen cycle. This includes both reactions that remove nitrogen from ecosystems (such as nitrate-based respiration and denitrification) as well as processes that introduce nitrogen (such as nitrate assimilation and nitrogen fixation).[181][182]
Researchers recently discovered archaeal involvement in ammonia oxidation reactions. These reactions are particularly important in the oceans.[183][184] In the sulfur cycle, archaea that grow by oxidizing sulfur compounds release this element from rocks, making it available to other organisms, but the archaea that do this, such as Sulfolobus, produce sulfuric acid as a waste product, and the growth of these organisms in abandoned mines can contribute to acid mine drainage and other environmental damage.[185] In the carbon cycle, methanogen archaea remove hydrogen and play an important role in the decay of organic matter by the populations of microorganisms that act as decomposers in anaerobic ecosystems, such as sediments and marshes.[186]
See also
- Bacterioplankton counting methods
- Bioluminescent bacteria
- Iron-oxidizing bacteria
- Pelagibacterales – model organisms in streamlining theory
- Streamlining theory
References
- ↑ Youngson, Robert M. (2006). Collins Dictionary of Human Biology. Glasgow: HarperCollins. ISBN 978-0-00-722134-9.
- ↑ Nelson, David L.; Cox, Michael M. (2005). Lehninger Principles of Biochemistry (4th ed.). New York: W.H. Freeman. ISBN 978-0-7167-4339-2. https://archive.org/details/lehningerprincip00lehn_0.
- ↑ Macmillan Dictionary of Life Sciences (2nd ed.). London: Macmillan Press. 1983. ISBN 978-0-333-34867-3.
- ↑ Fuerst, J. A. (2010). "Beyond Prokaryotes and Eukaryotes: Planctomycetes and Cell Organization". Nature Education 3 (9): 44. http://www.nature.com/scitable/topicpage/beyond-prokaryotes-and-eukaryotes-planctomycetes-and-cell-14158971.
- ↑ 2002 WHO mortality data Accessed 20 January 2007
- ↑ University of Georgia (10 December 2015). "Functions of global ocean microbiome key to understanding environmental changes". https://www.sciencedaily.com/releases/2015/12/151210181647.htm.
- ↑ "The biomass distribution on Earth". Proceedings of the National Academy of Sciences of the United States of America 115 (25): 6506–6511. 2018. doi:10.1073/pnas.1711842115. PMID 29784790. PMC 6016768. Bibcode: 1998PNAS...95.6578W. http://www.pnas.org/content/early/2018/05/15/1711842115.full.pdf.
- ↑ 8.0 8.1 Doglioni, Carlo; Pignatti, Johannes; Coleman, Max (2016). "Why did life develop on the surface of the Earth in the Cambrian?". Geoscience Frontiers 7 (6): 865–873. doi:10.1016/j.gsf.2016.02.001. Bibcode: 2016GeoFr...7..865D.
- ↑ "Age of the Earth". United States Geological Survey. 9 July 2007. http://pubs.usgs.gov/gip/geotime/age.html.
- ↑ Dalrymple 2001, pp. 205–221
- ↑ Manhesa, Gérard; Allègre, Claude J.; Dupréa, Bernard; Hamelin, Bruno (May 1980). "Lead isotope study of basic-ultrabasic layered complexes: Speculations about the age of the earth and primitive mantle characteristics". Earth and Planetary Science Letters 47 (3): 370–382. doi:10.1016/0012-821X(80)90024-2. Bibcode: 1980E&PSL..47..370M.
- ↑ Schopf, J. William; Kudryavtsev, Anatoliy B.; Czaja, Andrew D.; Tripathi, Abhishek B. (5 October 2007). "Evidence of Archean life: Stromatolites and microfossils". Precambrian Research 158 (3–4): 141–155. doi:10.1016/j.precamres.2007.04.009. Bibcode: 2007PreR..158..141S.
- ↑ Raven & Johnson 2002, p. 68
- ↑ Baumgartner, R. J. (2019). "Nano−porous pyrite and organic matter in 3.5-billion-year-old stromatolites record primordial life". Geology 47 (11): 1039–1043. doi:10.1130/G46365.1. Bibcode: 2019Geo....47.1039B. https://discovery.ucl.ac.uk/id/eprint/10087275/1/Baumgartner%20et%20al%202019%20accepted.pdf.
- ↑ Earliest signs of life: Scientists find microbial remains in ancient rocks Phys.org. 26 September 2019.
- ↑ Jablonski, David (25 June 1999). "The Future of the Fossil Record". Science 284 (5423): 2114–2116. doi:10.1126/science.284.5423.2114. PMID 10381868.
- ↑ 17.0 17.1 Cavalier-Smith, Thomas (29 June 2006). "Cell evolution and Earth history: stasis and revolution". Philosophical Transactions of the Royal Society B: Biological Sciences 361 (1470): 969–1006. doi:10.1098/rstb.2006.1842. PMID 16754610.
- ↑ Schopf, J. William (29 June 2006). "Fossil evidence of Archaean life". Philosophical Transactions of the Royal Society B: Biological Sciences 361 (1470): 869–885. doi:10.1098/rstb.2006.1834. PMID 16754604.
- Altermann, Wladyslaw; Kazmierczak, Józef (November 2003). "Archean microfossils: a reappraisal of early life on Earth". Research in Microbiology 154 (9): 611–617. doi:10.1016/j.resmic.2003.08.006. PMID 14596897.
- ↑ Schopf, J. William (19 July 1994). "Disparate rates, differing fates: tempo and mode of evolution changed from the Precambrian to the Phanerozoic". Proceedings of the National Academy of Sciences of the United States of America 91 (15): 6735–6742. doi:10.1073/pnas.91.15.6735. PMID 8041691. Bibcode: 1994PNAS...91.6735S.
- ↑ Poole, Anthony M.; Penny, David (January 2007). "Evaluating hypotheses for the origin of eukaryotes". BioEssays 29 (1): 74–84. doi:10.1002/bies.20516. PMID 17187354.
- ↑ 21.0 21.1 Dyall, Sabrina D.; Brown, Mark T.; Johnson, Patricia J. (9 April 2004). "Ancient Invasions: From Endosymbionts to Organelles". Science 304 (5668): 253–257. doi:10.1126/science.1094884. PMID 15073369. Bibcode: 2004Sci...304..253D.
- ↑ Martin, William (October 2005). "The missing link between hydrogenosomes and mitochondria". Trends in Microbiology 13 (10): 457–459. doi:10.1016/j.tim.2005.08.005. PMID 16109488.
- ↑ Lang, B. Franz; Gray, Michael W.; Burger, Gertraud (December 1999). "Mitochondrial genome evolution and the origin of eukaryotes". Annual Review of Genetics 33: 351–397. doi:10.1146/annurev.genet.33.1.351. PMID 10690412.
- McFadden, Geoffrey Ian (1 December 1999). "Endosymbiosis and evolution of the plant cell". Current Opinion in Plant Biology 2 (6): 513–519. doi:10.1016/S1369-5266(99)00025-4. PMID 10607659.
- ↑ Hug, Laura A.; Baker, Brett J.; Anantharaman, Karthik; Brown, Christopher T.; Probst, Alexander J.; Castelle, Cindy J.; Butterfield, Cristina N.; Hernsdorf, Alex W. et al. (2016-04-11). "A new view of the tree of life" (in en). Nature Microbiology 1 (5): 16048. doi:10.1038/nmicrobiol.2016.48. ISSN 2058-5276. PMID 27572647.
- ↑ Ciccarelli, Francesca D.; Doerks, Tobias; von Mering, Christian et al. (3 March 2006). "Toward Automatic Reconstruction of a Highly Resolved Tree of Life". Science 311 (5765): 1283–1287. doi:10.1126/science.1123061. PMID 16513982. Bibcode: 2006Sci...311.1283C.
- ↑ DeLong, Edward F.; Pace, Norman R. (1 August 2001). "Environmental Diversity of Bacteria and Archaea". Systematic Biology 50 (4): 470–478. doi:10.1080/106351501750435040. PMID 12116647.
- ↑ Kaiser, Dale (December 2001). "Building a multicellular organism". Annual Review of Genetics 35: 103–123. doi:10.1146/annurev.genet.35.102401.090145. PMID 11700279.
- ↑ Zimmer, Carl (7 January 2016). "Genetic Flip Helped Organisms Go From One Cell to Many". The New York Times. https://www.nytimes.com/2016/01/12/science/genetic-flip-helped-organisms-go-from-one-cell-to-many.html.
- ↑ Valentine, James W.; Jablonski, David; Erwin, Douglas H. (1 March 1999). "Fossils, molecules and embryos: new perspectives on the Cambrian explosion". Development 126 (5): 851–859. doi:10.1242/dev.126.5.851. PMID 9927587. http://dev.biologists.org/content/126/5/851.full.pdf+html. Retrieved 2014-12-30.
- ↑ Ohno, Susumu (January 1997). "The reason for as well as the consequence of the Cambrian explosion in animal evolution". Journal of Molecular Evolution 44 (Suppl. 1): S23–S27. doi:10.1007/PL00000055. PMID 9071008. Bibcode: 1997JMolE..44S..23O.
- Valentine, James W.; Jablonski, David (2003). "Morphological and developmental macroevolution: a paleontological perspective". The International Journal of Developmental Biology 47 (7–8): 517–522. PMID 14756327. http://www.ijdb.ehu.es/web/paper.php?doi=14756327. Retrieved 2014-12-30.
- ↑ Campbell, N. "Biology:Concepts & Connections". Pearson Education. San Francisco: 2003.
- ↑ Harper, Douglas. "prokaryote". Online Etymology Dictionary. https://www.etymonline.com/?term=prokaryote.
- ↑ Harper, Douglas. "eukaryotic". Online Etymology Dictionary. https://www.etymonline.com/?term=eukaryotic.
- ↑ "The concept of a bacterium". Archiv für Mikrobiologie 42: 17–35. 1962. doi:10.1007/BF00425185. PMID 13916221.
- ↑ "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proceedings of the National Academy of Sciences of the United States of America 74 (11): 5088–90. November 1977. doi:10.1073/pnas.74.11.5088. PMID 270744. Bibcode: 1977PNAS...74.5088W.
- ↑ "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America 87 (12): 4576–9. June 1990. doi:10.1073/pnas.87.12.4576. PMID 2112744. Bibcode: 1990PNAS...87.4576W.
- ↑ Ohtomo, Yoko; Kakegawa, Takeshi; Ishida, Akizumi; Nagase, Toshiro; Rosing, Minik T. (January 2014). "Evidence for biogenic graphite in early Archaean Isua metasedimentary rocks" (in En). Nature Geoscience 7 (1): 25–28. doi:10.1038/ngeo2025. Bibcode: 2014NatGe...7...25O.
- ↑ Nutman, Allen P.; Bennett, Vickie C.; Friend, Clark R. L.; Kranendonk, Martin J. Van; Chivas, Allan R. (September 2016). "Rapid emergence of life shown by discovery of 3,700-million-year-old microbial structures" (in En). Nature 537 (7621): 535–538. doi:10.1038/nature19355. PMID 27580034. Bibcode: 2016Natur.537..535N. http://ro.uow.edu.au/cgi/viewcontent.cgi?article=5181&context=smhpapers.
- ↑ Borenstein, Seth (19 October 2015). "Hints of life on what was thought to be desolate early Earth". Excite. Associated Press (Yonkers, NY: Mindspark Interactive Network). http://apnews.excite.com/article/20151019/us-sci--earliest_life-a400435d0d.html.
- ↑ Bell, Elizabeth A.; Boehnike, Patrick; Harrison, T. Mark et al. (19 October 2015). "Potentially biogenic carbon preserved in a 4.1 billion-year-old zircon". Proc. Natl. Acad. Sci. U.S.A. 112 (47): 14518–21. doi:10.1073/pnas.1517557112. PMID 26483481. Bibcode: 2015PNAS..11214518B.
- ↑ Dodd, Matthew S.; Papineau, Dominic; Grenne, Tor; slack, John F.; Rittner, Martin; Pirajno, Franco; O'Neil, Jonathan; Little, Crispin T. S. (2 March 2017). "Evidence for early life in Earth's oldest hydrothermal vent precipitates". Nature 543 (7643): 60–64. doi:10.1038/nature21377. PMID 28252057. Bibcode: 2017Natur.543...60D. http://eprints.whiterose.ac.uk/112179/1/ppnature21377_Dodd_for%20Symplectic.pdf.
- ↑ Zimmer, Carl (1 March 2017). "Scientists Say Canadian Bacteria Fossils May Be Earth's Oldest". The New York Times. https://www.nytimes.com/2017/03/01/science/earths-oldest-bacteria-fossils.html.
- ↑ Nisbet, Euan G.; Fowler, C. M. R. (December 7, 1999). "Archaean metabolic evolution of microbial mats". Proceedings of the Royal Society of London B 266 (1436): 2375–2382. doi:10.1098/rspb.1999.0934.
- ↑ Anbar, Ariel D.; Yun Duan; Lyons, Timothy W. et al. (September 28, 2007). "A Whiff of Oxygen Before the Great Oxidation Event?". Science 317 (5846): 1903–1906. doi:10.1126/science.1140325. PMID 17901330. Bibcode: 2007Sci...317.1903A.
- ↑ Knoll, Andrew H.; Javaux, Emmanuelle J.; Hewitt, David; Cohen, Phoebe (June 29, 2006). "Eukaryotic organisms in Proterozoic oceans". Philosophical Transactions of the Royal Society B 361 (1470): 1023–1038. doi:10.1098/rstb.2006.1843. PMID 16754612.
- ↑ Fedonkin, Mikhail A. (March 31, 2003). "The origin of the Metazoa in the light of the Proterozoic fossil record". Paleontological Research 7 (1): 9–41. doi:10.2517/prpsj.7.9. http://www.vend.paleo.ru/pub/Fedonkin_2003.pdf. Retrieved 2008-09-02.
- ↑ Bonner, John Tyler (1998). "The origins of multicellularity". Integrative Biology 1 (1): 27–36. doi:10.1002/(SICI)1520-6602(1998)1:1<27::AID-INBI4>3.0.CO;2-6.
- ↑ May, R.M. (1988) "How many species are there on earth?". Science, 241(4872): 1441–1449. doi:10.1126/science.241.4872.1441.
- ↑ Locey, K.J. and Lennon, J.T. (2016) "Scaling laws predict global microbial diversity". Proceedings of the National Academy of Sciences, 113(21): 5970–5975. doi:10.1073/pnas.1521291113.
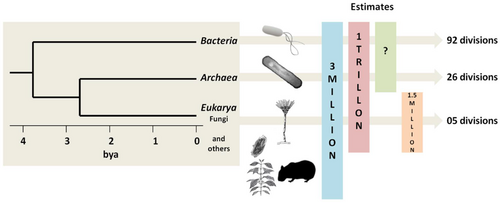
- ↑ Vitorino, L.C. and Bessa, L.A. (2018) "Microbial diversity: the gap between the estimated and the known". Diversity, 10(2): 46. doi:10.3390/d10020046. 50px Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Living Bacteria Are Riding Earth’s Air Currents Smithsonian Magazine, 11 January 2016.
- ↑ Robbins, Jim (13 April 2018). "Trillions Upon Trillions of Viruses Fall From the Sky Each Day". The New York Times. https://www.nytimes.com/2018/04/13/science/virosphere-evolution.html.
- ↑ Reche, Isabel; D’Orta, Gaetano; Mladenov, Natalie; Winget, Danielle M; Suttle, Curtis A (29 January 2018). "Deposition rates of viruses and bacteria above the atmospheric boundary layer". ISME Journal 12 (4): 1154–1162. doi:10.1038/s41396-017-0042-4. PMID 29379178.
- ↑ Suttle, C.A. (2005). "Viruses in the Sea". Nature 437 (9): 356–361. doi:10.1038/nature04160. PMID 16163346. Bibcode: 2005Natur.437..356S.
- ↑ Shors 2017, p. 5
- ↑ Shors 2017, p. 593
- ↑ Suttle CA (2007). "Marine viruses—major players in the global ecosystem". Nature Reviews Microbiology 5 (10): 801–12. doi:10.1038/nrmicro1750. PMID 17853907.
- ↑ Schulz, Kestin; Smit, Mariya W.; Herfort, Lydie; Simon, Holly M. (2018). "The Unseen World in the River". Frontiers for Young Minds 6. doi:10.3389/frym.2018.00004. 50px Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ "Walsby's square bacterium: fine structure of an orthogonal procaryote". Journal of Bacteriology 148 (1): 352–60. October 1981. doi:10.1128/JB.148.1.352-360.1981. PMID 7287626.
- ↑ Durham, Bryndan P.; Grote, Jana; Whittaker, Kerry A.; Bender, Sara J.; Luo, Haiwei; Grim, Sharon L.; Brown, Julia M.; Casey, John R. et al. (2014). "Draft genome sequence of marine alphaproteobacterial strain HIMB11, the first cultivated representative of a unique lineage within the Roseobacter clade possessing an unusually small genome". Standards in Genomic Sciences 9 (3): 632–645. doi:10.4056/sigs.4998989. PMID 25197450.
- ↑ "Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington state". Applied and Environmental Microbiology 70 (7): 4230–41. 2004. doi:10.1128/AEM.70.7.4230-4241.2004. PMID 15240306. Bibcode: 2004ApEnM..70.4230F.
- ↑ "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America 87 (12): 4576–9. 1990. doi:10.1073/pnas.87.12.4576. PMID 2112744. Bibcode: 1990PNAS...87.4576W.
- ↑ "Disparate rates, differing fates: tempo and mode of evolution changed from the Precambrian to the Phanerozoic". Proceedings of the National Academy of Sciences of the United States of America 91 (15): 6735–42. 1994. doi:10.1073/pnas.91.15.6735. PMID 8041691. Bibcode: 1994PNAS...91.6735S.
- ↑ "Environmental diversity of bacteria and archaea". Systematic Biology 50 (4): 470–8. 2001. doi:10.1080/106351501750435040. PMID 12116647.
- ↑ "Archaea and the prokaryote-to-eukaryote transition". Microbiology and Molecular Biology Reviews 61 (4): 456–502. 1997. doi:10.1128/mmbr.61.4.456-502.1997. PMID 9409149.
- ↑ "Evaluating hypotheses for the origin of eukaryotes". BioEssays 29 (1): 74–84. 2007. doi:10.1002/bies.20516. PMID 17187354.
- ↑ "Mitochondrial genome evolution and the origin of eukaryotes". Annual Review of Genetics 33: 351–97. 1999. doi:10.1146/annurev.genet.33.1.351. PMID 10690412.
- ↑ "Endosymbiosis and evolution of the plant cell". Current Opinion in Plant Biology 2 (6): 513–9. 1999. doi:10.1016/S1369-5266(99)00025-4. PMID 10607659.
- ↑ "Control of the bacterial cell cycle by cytoplasmic growth". Critical Reviews in Microbiology 28 (1): 61–77. 2002. doi:10.1080/1040-840291046696. PMID 12003041.
- ↑ "Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes". Journal of Bacteriology 83 (4): 736–37. April 1962. doi:10.1128/jb.83.4.736-737.1962. PMID 13888946.
- ↑ "Candidatus Pelagibacter Ubique." European Bioinformatics Institute. European Bioinformatics Institute, 2011. Web. 08 Jan. 2012. http://www.ebi.ac.uk/2can/genomes/bacteria/Candidatus_Pelagibacter_ubique.html
- ↑ "Flea market: A newly discovered virus may be the most abundant organism on the planet". The Economist. 16 February 2013. https://www.economist.com/news/science-and-technology/21571843-newly-discovered-virus-may-be-most-abundant-organism-planet-flea.
- ↑ Zhao, Y.; Temperton, B.; Thrash, J. C.; Schwalbach, M. S.; Vergin, K. L.; Landry, Z. C.; Ellisman, M.; Deerinck, T. et al. (2013). "Abundant SAR11 viruses in the ocean". Nature 494 (7437): 357–360. doi:10.1038/nature11921. PMID 23407494. Bibcode: 2013Natur.494..357Z.
- ↑ Bentzon-Tilia, Mikkel; Gram, Lone (2017). "Biotechnological Applications of the Roseobacter Clade". Bioprospecting. Topics in Biodiversity and Conservation. 16. Springer, Cham. pp. 137–166. doi:10.1007/978-3-319-47935-4_7. ISBN 978-3-319-47933-0.
- ↑ NCBI Taxonomy Browser: Roseobacter National Center for Biotechnology Information. Accessed: 8 May 2020.
- ↑ "The largest Bacterium: Scientist discovers new bacterial life form off the African coast", Max Planck Institute for Marine Microbiology, 8 April 1999, http://www.mpg.de/english/illustrationsDocumentation/documentation/pressReleases/1999/news17_99.htm
- ↑ List of Prokaryotic names with Standing in Nomenclature - Genus Thiomargarita, https://lpsn.dsmz.de/genus/thiomargarita
- ↑ Changes in oxygen concentrations in our ocean can disrupt fundamental biological cycles Phys.org, 25 November 2019.
- ↑ "Cyanobacteria and cyanobacterial toxins". Oceans and Human Health: Risks and Remedies from the Seas. Academic Press. 2 September 2011. pp. 271–296. ISBN 978-0-08-087782-2. https://books.google.com/books?id=LMZPqW-PmFYC&pg=PA271.
- ↑ "The Rise of Oxygen - Astrobiology Magazine" (in en-US). 30 July 2003. http://www.astrobio.net/news-exclusive/the-rise-of-oxygen/.
- ↑ Flannery, D. T.; R.M. Walter (2012). "Archean tufted microbial mats and the Great Oxidation Event: new insights into an ancient problem". Australian Journal of Earth Sciences 59 (1): 1–11. doi:10.1080/08120099.2011.607849. Bibcode: 2012AuJES..59....1F.
- ↑ Rothschild, Lynn (September 2003). "Understand the evolutionary mechanisms and environmental limits of life". NASA. http://astrobiology.arc.nasa.gov/roadmap/g5.html.
- ↑ Basu, Subhajit; Gledhill, Martha; De Beer, Dirk; Prabhu Matondkar, S. G.; Shaked, Yeala (2019). "Colonies of marine cyanobacteria Trichodesmium interact with associated bacteria to acquire iron from dust". Communications Biology 2: 284. doi:10.1038/s42003-019-0534-z. PMID 31396564. 50px Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Patrick J. Keeling (2004). "Diversity and evolutionary history of plastids and their hosts". American Journal of Botany 91 (10): 1481–1493. doi:10.3732/ajb.91.10.1481. PMID 21652304.
- ↑ "The cells that rule the seas". Scientific American 289 (6): 52–3. December 2003. doi:10.1038/scientificamerican1203-52. PMID 14631732. Bibcode: 2003SciAm.289f..52N. http://guowei.ccps.tp.edu.tw/nc/UploadDocument/255_02%20The%20Cells%20that%20Rules%20the%20Sea.pdf. Retrieved 2 June 2019.
- ↑ "The Most Important Microbe You've Never Heard Of". https://www.npr.org/templates/story/story.php?storyId=91448837.
- ↑ Flombaum, P.; Gallegos, J. L.; Gordillo, R. A.; Rincon, J.; Zabala, L. L.; Jiao, N.; Karl, D. M.; Li, W. K. W. et al. (2013). "Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus". Proceedings of the National Academy of Sciences 110 (24): 9824–9829. doi:10.1073/pnas.1307701110. PMID 23703908. Bibcode: 2013PNAS..110.9824F.
- ↑ Nabors, Murray W. (2004). Introduction to Botany. San Francisco, CA: Pearson Education, Inc. ISBN 978-0-8053-4416-5.
- ↑ Allaby, M., ed (1992). "Algae". The Concise Dictionary of Botany. Oxford: Oxford University Press.
- ↑ Ivars-Martínez, Elena; d'Auria, Giuseppe; Rodríguez-Valera, Francisco; Sánchez-Porro, Cristina; Ventosa, Antonio; Joint, IAN; Mühling, Martin (2008). "Biogeography of the ubiquitous marine bacterium Alteromonas macleodiidetermined by multilocus sequence analysis". Molecular Ecology 17 (18): 4092–4106. doi:10.1111/j.1365-294x.2008.03883.x. PMID 19238708.
- ↑ López-Pérez, Mario; Gonzaga, Aitor; Martin-Cuadrado, Ana-Belen; Onyshchenko, Olga; Ghavidel, Akbar; Ghai, Rohit; Rodriguez-Valera, Francisco (2012). "Genomes of surface isolates of Alteromonas macleodii: The life of a widespread marine opportunistic copiotroph". Scientific Reports 2: 696. doi:10.1038/srep00696. PMID 23019517. Bibcode: 2012NatSR...2E.696L.
- ↑ "Archaea associated with human surfaces: not to be underestimated". FEMS Microbiology Reviews 39 (5): 631–48. 2015. doi:10.1093/femsre/fuv010. PMID 25907112.
- ↑ Archaea Online Etymology Dictionary. Retrieved 17 August 2016.
- ↑ Pace NR (May 2006). "Time for a change". Nature 441 (7091): 289. doi:10.1038/441289a. PMID 16710401. Bibcode: 2006Natur.441..289P.
- ↑ Stoeckenius W (1 October 1981). "Walsby's square bacterium: fine structure of an orthogonal procaryote". Journal of Bacteriology 148 (1): 352–60. doi:10.1128/JB.148.1.352-360.1981. PMID 7287626.
- ↑ Brock Biology of Microorganisms (11th ed.). Prentice Hall. 2005. ISBN 978-0-13-144329-7.
- ↑ Huber, Harald (2002). "A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont". Nature 417 (6884): 63–67. doi:10.1038/417063a. PMID 11986665. Bibcode: 2002Natur.417...63H.
- ↑ Brochier, Celine; Gribaldo, S; Zivanovic, Y; Confalonieri, F et al. (2005). "Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales?". Genome Biology 6 (5): R42. doi:10.1186/gb-2005-6-5-r42. PMID 15892870.
- ↑ Orellana, Luis H.; Ben Francis, T.; Krüger, Karen; Teeling, Hanno; Müller, Marie-Caroline; Fuchs, Bernhard M.; Konstantinidis, Konstantinos T.; Amann, Rudolf I. (2019). "Niche differentiation among annually recurrent coastal Marine Group II Euryarchaeota". The ISME Journal 13 (12): 3024–3036. doi:10.1038/s41396-019-0491-z. PMID 31447484.
- ↑ See especially Fig. 4 in Nishimura, Yosuke; Watai, Hiroyasu; Honda, Takashi; Mihara, Tomoko; Omae, Kimiho; Roux, Simon; Blanc-Mathieu, Romain; Yamamoto, Keigo et al. (2017). "Environmental Viral Genomes Shed New Light on Virus-Host Interactions in the Ocean". mSphere 2 (2). doi:10.1128/mSphere.00359-16. PMID 28261669.
- ↑ Philosof, Alon; Yutin, Natalya; Flores-Uribe, José; Sharon, Itai; Koonin, Eugene V.; Béjà, Oded (2017). "Novel Abundant Oceanic Viruses of Uncultured Marine Group II Euryarchaeota". Current Biology 27 (9): 1362–1368. doi:10.1016/j.cub.2017.03.052. PMID 28457865.
- ↑ Xia, Xiaomin; Guo, Wang; Liu, Hongbin (2017). "Basin Scale Variation on the Composition and Diversity of Archaea in the Pacific Ocean". Frontiers in Microbiology 8: 2057. doi:10.3389/fmicb.2017.02057. PMID 29109713.
- ↑ Martin-Cuadrado, Ana-Belen; Garcia-Heredia, Inmaculada; Moltó, Aitor Gonzaga; López-Úbeda, Rebeca; Kimes, Nikole; López-García, Purificación; Moreira, David; Rodriguez-Valera, Francisco (2015). "A new class of marine Euryarchaeota group II from the mediterranean deep chlorophyll maximum". The ISME Journal 9 (7): 1619–1634. doi:10.1038/ismej.2014.249. PMID 25535935.
- ↑ NCBI: Candidatus Poseidoniales (order)
- ↑ NCBI: Marine Group III
- ↑ NCBI: Marine Group IV
- ↑ "Comparative biochemistry of Archaea and Bacteria". Current Opinion in Genetics & Development 1 (4): 544–51. December 1991. doi:10.1016/S0959-437X(05)80206-0. PMID 1822288.
- ↑ Microbiology: An Evolving Science (3 ed.). WW Norton & Company. pp. 491–44.
- ↑ "Extremophile". Encyclopedia of Earth. National Council of Science & the Environment. 2010. http://www.eoearth.org/article/Extremophile?topic=49540.
- ↑ Yurkov, V. V.; Beatty, J. T. (1998). "Aerobic anoxygenic phototrophic bacteria". Microbiology and Molecular Biology Reviews 62 (3): 695–724. doi:10.1128/MMBR.62.3.695-724.1998. PMID 9729607.
- ↑ Lami, R.; Cottrell, M. T.; Ras, J.; Ulloa, O.; Obernosterer, I.; Claustre, H.; Kirchman, D. L.; Lebaron, P. (2007). "High Abundances of Aerobic Anoxygenic Photosynthetic Bacteria in the South Pacific Ocean". Applied and Environmental Microbiology 73 (13): 4198–205. doi:10.1128/AEM.02652-06. PMID 17496136. Bibcode: 2007ApEnM..73.4198L.
- ↑ Life science weekly. (2012). Bacteria; Reports from Spanish National Research Council (CSIC) Describe Recent Advances in Bacteria. ISSN 1552-2466. P.4582.
- ↑ Jiao, Nianzhi; Zhang, Yao; Zeng, Yonghui; Hong, Ning; Liu, Rulong; Chen, Feng; Wang, Pinxian (2007). "Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean". Environmental Microbiology 9 (12): 3091–3099. doi:10.1111/j.1462-2920.2007.01419.x. PMID 17991036.
- ↑ Lami, Raphaël; Cottrell, Matthew T.; Ras, JoséPhine; Ulloa, Osvaldo; Obernosterer, Ingrid; Claustre, Hervé; Kirchman, David L.; Lebaron, Philippe (2007). "High Abundances of Aerobic Anoxygenic Photosynthetic Bacteria in the South Pacific Ocean". Applied and Environmental Microbiology 73 (13): 4198–4205. doi:10.1128/AEM.02652-06. PMID 17496136. Bibcode: 2007ApEnM..73.4198L.
- ↑ Kolber, Z. S. (2001). "Contribution of Aerobic Photoheterotrophic Bacteria to the Carbon Cycle in the Ocean". Science 292 (5526): 2492–2495. doi:10.1126/science.1059707. PMID 11431568.
- ↑ Aragno, Michel; Schlegel, Hans G. (1981). "The Hydrogen-Oxidizing Bacteria". in Starr, Mortimer P.; Stolp, Heinz; Trüper, Hans G. et al.. The Prokaryotes. Berlin, Heidelberg: Springer. pp. 865–893. doi:10.1007/978-3-662-13187-9_70. ISBN 978-3-662-13187-9.
- ↑ 117.0 117.1 "The archaellum: how Archaea swim". Frontiers in Microbiology 6: 23. 27 January 2015. doi:10.3389/fmicb.2015.00023. PMID 25699024.
- ↑ "Flagellar rotation and the mechanism of bacterial motility". Nature 249 (452): 73–4. May 1974. doi:10.1038/249073a0. PMID 4598030. Bibcode: 1974Natur.249...73S.
- ↑ "Rapid rotation of flagellar bundles in swimming bacteria". Nature 325 (6105): 637–640. 1987. doi:10.1038/325637a0. Bibcode: 1987Natur.325..637L.
- ↑ "Bacteria swim by rotating their flagellar filaments". Nature 245 (5425): 380–2. October 1973. doi:10.1038/245380a0. PMID 4593496. Bibcode: 1973Natur.245..380B.
- ↑ "Movement and locomotion of microorganisms". Annual Review of Microbiology 19: 21–58. 1965. doi:10.1146/annurev.mi.19.100165.000321. PMID 5318439.
- ↑ "Bacterial motility on a surface: many ways to a common goal". Annual Review of Microbiology 57: 249–73. 2003. doi:10.1146/annurev.micro.57.030502.091014. PMID 14527279.
- ↑ "Archaeal Flagella and Pili". Pili and Flagella: Current Research and Future Trends. Caister Academic Press. 2009. ISBN 978-1-904455-48-6.
- ↑ Brady, Richard M. (1993). "Torque and switching in the bacterial flagellar motor. An electrostatic model". Biophysical Journal 64 (4): 961–973. doi:10.1016/S0006-3495(93)81462-0. PMID 7684268. Bibcode: 1993BpJ....64..961B.
- ↑ "Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP". Journal of Molecular Biology 384 (1): 1–8. 2008. doi:10.1016/j.jmb.2008.08.057. PMID 18786541.
- ↑ 126.0 126.1 Skerker, J. M.; Berg, H. C. (2001-06-05). "Direct observation of extension and retraction of type IV pili". Proceedings of the National Academy of Sciences of the United States of America 98 (12): 6901–6904. doi:10.1073/pnas.121171698. ISSN 0027-8424. PMID 11381130. Bibcode: 2001PNAS...98.6901S.
- ↑ Mattick, John S. (2002). "Type IV pili and twitching motility". Annual Review of Microbiology 56: 289–314. doi:10.1146/annurev.micro.56.012302.160938. ISSN 0066-4227. PMID 12142488.
- ↑ Merz, A. J.; So, M.; Sheetz, M. P. (2000-09-07). "Pilus retraction powers bacterial twitching motility". Nature 407 (6800): 98–102. doi:10.1038/35024105. ISSN 0028-0836. PMID 10993081. Bibcode: 2000Natur.407...98M.
- ↑ Henrichsen, J. (December 1972). "Bacterial surface translocation: a survey and a classification". Bacteriological Reviews 36 (4): 478–503. doi:10.1128/BR.36.4.478-503.1972. ISSN 0005-3678. PMID 4631369.
- ↑ Nan, Beiyan (February 2017). "Bacterial Gliding Motility: Rolling Out a Consensus Model". Current Biology 27 (4): R154–R156. doi:10.1016/j.cub.2016.12.035. PMID 28222296.
- ↑ Nan, Beiyan; McBride, Mark J.; Chen, Jing; Zusman, David R.; Oster, George (February 2014). "Bacteria that Glide with Helical Tracks". Current Biology 24 (4): 169–174. doi:10.1016/j.cub.2013.12.034. PMID 24556443.
- ↑ Sibley, L.David; Håkansson, Sebastian; Carruthers, Vern B (1998-01-01). "Gliding motility: An efficient mechanism for cell penetration". Current Biology 8 (1): R12–R14. doi:10.1016/S0960-9822(98)70008-9. PMID 9427622.
- ↑ Sibley, LDI (Oct 2010). "How apicomplexan parasites move in and out of cells". Curr Opin Biotechnol 21 (5): 592–8. doi:10.1016/j.copbio.2010.05.009. PMID 20580218.
- ↑ Harshey, Rasika M. (2003-01-01). "Bacterial Motility on a Surface: Many Ways to a Common Goal". Annual Review of Microbiology 57 (1): 249–73. doi:10.1146/annurev.micro.57.030502.091014. PMID 14527279.
- ↑ Henrichsen, J (1972). "Bacterial surface translocation: a survey and a classification". Bacteriological Reviews 36 (4): 478–503. doi:10.1128/BR.36.4.478-503.1972. PMID 4631369.
- ↑ "BIOL 230 Lab Manual: Nonmotile Bacteria in Motility Medium". http://faculty.ccbcmd.edu/courses/bio141/labmanua/lab7/nonmotil.html.
- ↑ Pósfai, M., Lefèvre, C., Trubitsyn, D., Bazylinski, D.A. and Frankel, R. (2013) "Phylogenetic significance of composition and crystal morphology of magnetosome minerals". Frontiers in microbiology, 4: 344. doi:10.3389/fmicb.2013.00344.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 3.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 3.0 International License.
- ↑ Lin, Wei; Zhang, Wensi; Zhao, Xiang; Roberts, Andrew; Paterson, Greig; Bazylinski, Dennis; Pan, Yongxin (March 2018). "Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution". The ISME Journal 12 (6): 1508–1519. doi:10.1038/s41396-018-0098-9. PMID 29581530.
- ↑ 139.0 139.1 Dusenbery, David B. (2009). Living at micro scale : the unexpected physics of being small. Cambridge, Mass.: Harvard University Press. pp. 100–101. ISBN 978-0-674-03116-6.
- ↑ 140.0 140.1 Zhang, W.J. and Wu, L.F., 2020. Flagella and Swimming Behavior of Marine Magnetotactic Bacteria. Biomolecules, 10(3), p.460. doi:10.3390/biom10030460.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Jogler, C.; Wanner, G.; Kolinko, S.; Niebler, M.; Amann, R.; Petersen, N.; Kube, M.; Reinhardt, R. et al. (2010). "Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep-branching Nitrospira phylum". Proceedings of the National Academy of Sciences 108 (3): 1134–1139. doi:10.1073/pnas.1012694108. PMID 21191098. Bibcode: 2011PNAS..108.1134J.
- ↑ Monteil, Caroline L.; Vallenet, David; Menguy, Nicolas; Benzerara, Karim; Barbe, Valérie; Fouteau, Stéphanie; Cruaud, Corinne; Floriani, Magali et al. (July 2019). "Ectosymbiotic bacteria at the origin of magnetoreception in a marine protist" (in en). Nature Microbiology 4 (7): 1088–1095. doi:10.1038/s41564-019-0432-7. ISSN 2058-5276. PMID 31036911.
- ↑ Natan, Eviatar; Fitak, Robert Rodgers; Werber, Yuval; Vortman, Yoni (2020-09-28). "Symbiotic magnetic sensing: raising evidence and beyond". Philosophical Transactions of the Royal Society B: Biological Sciences 375 (1808): 20190595. doi:10.1098/rstb.2019.0595. PMID 32772668.
- ↑ Kopp, R. E.; Kirschvink, J. L. (2007). "The identification and biogeochemical interpretation of fossil magnetotactic bacteria". Earth-Science Reviews 86 (1–4): 42–61. doi:10.1016/j.earscirev.2007.08.001. Bibcode: 2008ESRv...86...42K. https://authors.library.caltech.edu/9352/1/Kopp2008_Postprint_The_identification_and_interpretation_of_fossil_magnetotactic_bacteria.pdf.
- ↑ Chang, S. R.; J. L. Kirschvink (1989). "Magnetofossils, the magnetization of sediments, and the evolution of magnetite biomineralization". Annual Review of Earth and Planetary Sciences 17: 169–195. doi:10.1146/annurev.ea.17.050189.001125. Bibcode: 1989AREPS..17..169C.
- ↑ Walsby AE (1969). "The Permeability of Blue-Green Algal Gas-Vacuole Membranes to Gas". Proceedings of the Royal Society of London. Series B, Biological Sciences 173 (1031): 235–255. doi:10.1098/rspb.1969.0049. OCLC 479422015. Bibcode: 1969RSPSB.173..235W.
- ↑ "Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation". Applied and Environmental Microbiology 70 (12): 7487–96. December 2004. doi:10.1128/AEM.70.12.7487-7496.2004. PMID 15574952. Bibcode: 2004ApEnM..70.7487K.
- ↑ Schulz-Vogt, Heide N. (2006). "Vacuoles". Inclusions in Prokaryotes. Microbiology Monographs. 1. pp. 295–298. doi:10.1007/3-540-33774-1_10. ISBN 978-3-540-26205-3.
- ↑ Montánchez, Itxaso; Ogayar, Elixabet; Plágaro, Ander Hernández; Esteve-Codina, Anna; Gómez-Garrido, Jèssica; Orruño, Maite; Arana, Inés; Kaberdin, Vladimir R. (2019). "Analysis of Vibrio harveyi adaptation in sea water microcosms at elevated temperature provides insights into the putative mechanisms of its persistence and spread in the time of global warming". Scientific Reports 9 (1): 289. doi:10.1038/s41598-018-36483-0. PMID 30670759. Bibcode: 2019NatSR...9..289M.
- ↑ McFall-Ngai, Margaret; Heath-Heckman, Elizabeth A.C.; Gillette, Amani A.; Peyer, Suzanne M.; Harvie, Elizabeth A. (2012). "The secret languages of coevolved symbioses: Insights from the Euprymna scolopes–Vibrio fischeri symbiosis". Seminars in Immunology 24 (1): 3–8. doi:10.1016/j.smim.2011.11.006. PMID 22154556.
- ↑ Waters, Christopher M.; Bassler, Bonnie L. (2005-10-07). "QUORUM SENSING: Cell-to-Cell Communication in Bacteria" (in en). Annual Review of Cell and Developmental Biology 21 (1): 319–346. doi:10.1146/annurev.cellbio.21.012704.131001. PMID 16212498.
- ↑ Young, R.; Roper, C. (1976). "Bioluminescent countershading in midwater animals: Evidence from living squid". Science 191 (4231): 1046–1048. doi:10.1126/science.1251214. PMID 1251214. Bibcode: 1976Sci...191.1046Y.
- ↑ Owens, Leigh; Busico-Salcedo, Nancy (2006). "Vibrio harveyi: Pretty Problems in Paradise (Chapter 19)". in Thompson, Fabiano; Austin, Brian; Swings, Jean. The Biology of Vibrios. ASM Press.
- ↑ DeLong, E.F.; Beja, O. (2010). "The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times". PLOS Biology 8 (4): e1000359. doi:10.1371/journal.pbio.1000359. e1000359. PMID 20436957.
- ↑ 155.0 155.1 Gómez-Consarnau, L.; Raven, J.A.; Levine, N.M.; Cutter, L.S.; Wang, D.; Seegers, B.; Arístegui, J.; Fuhrman, J.A. et al. (2019). "Microbial rhodopsins are major contributors to the solar energy captured in the sea". Science Advances 5 (8): eaaw8855. doi:10.1126/sciadv.aaw8855. PMID 31457093. Bibcode: 2019SciA....5.8855G.
- ↑ Oren, Aharon (2002). "Molecular ecology of extremely halophilic Archaea and Bacteria". FEMS Microbiology Ecology 39 (1): 1–7. doi:10.1111/j.1574-6941.2002.tb00900.x. PMID 19709178.
- ↑ Béja, O.; Aravind, L.; Koonin, E.V.; Suzuki, M.T.; Hadd, A.; Nguyen, L.P.; Jovanovich, S.B.; Gates, C.M. et al. (2000). "Bacterial rhodopsin: evidence for a new type of phototrophy in the sea". Science 289 (5486): 1902–1906. doi:10.1126/science.289.5486.1902. PMID 10988064. Bibcode: 2000Sci...289.1902B.
- ↑ "Interviews with Fellows: Ed Delong". American Academy of Microbiology. http://academy.asm.org/index.php/news-views/interviews-with-fellows/366-ed-delong.
- ↑ Bacteria with Batteries, Popular Science, January 2001, Page 55.
- ↑ Béja, O.; Aravind, L.; Koonin, E.V.; Suzuki, M.T.; Hadd, A.; Nguyen, L.P.; Jovanovich, S.B.; Gates, C.M. et al. (2000). "Bacterial rhodopsin: evidence for a new type of phototrophy in the sea". Science 289 (5486): 1902–1906. doi:10.1126/science.289.5486.1902. PMID 10988064. Bibcode: 2000Sci...289.1902B.
- ↑ Boeuf, Dominique; Audic, Stéphane; Brillet-Guéguen, Loraine; Caron, Christophe; Jeanthon, Christian (2015). "MicRhoDE: a curated database for the analysis of microbial rhodopsin diversity and evolution". Database 2015: bav080. doi:10.1093/database/bav080. PMID 26286928.
- ↑ Yawo, Hiromu; Kandori, Hideki; Koizumi, Amane (5 June 2015). Optogenetics: Light-Sensing Proteins and Their Applications. Springer. pp. 3–4. ISBN 978-4-431-55516-2. https://books.google.com/books?id=5M3WCQAAQBAJ&pg=PA3. Retrieved 30 September 2015.
- ↑ A tiny marine microbe could play a big role in climate change University of Southern California, Press Room, 8 August 2019.
- ↑ DasSarma, Shiladitya; Schwieterman, Edward W. (11 October 2018). "Early evolution of purple retinal pigments on Earth and implications for exoplanet biosignatures". International Journal of Astrobiology 20 (3): 241–250. doi:10.1017/S1473550418000423. ISSN 1473-5504.
- ↑ Sparks, William B.; DasSarma, S.; Reid, I. N. (December 2006). "Evolutionary Competition Between Primitive Photosynthetic Systems: Existence of an early purple Earth?". American Astronomical Society Meeting Abstracts 38: 901. Bibcode: 2006AAS...209.0605S.
- ↑ Dane Konop (July 29, 1997). "Scientists discover methane ice worms on Gulf of Mexico sea floor". National Oceanic and Atmospheric Administration. http://www.publicaffairs.noaa.gov/pr97/jul97/noaa97-44.html.
- ↑ Sharp, Koty H.; Pratte, Zoe A.; Kerwin, Allison H.; Rotjan, Randi D.; Stewart, Frank J. (2017). "Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata". Microbiome 5 (1): 120. doi:10.1186/s40168-017-0329-8. PMID 28915923.
- ↑ Lema, K.A., Willis, B.L. and Bourne, D.G. (2012) "Corals form characteristic associations with symbiotic nitrogen-fixing bacteria". Applied and Environmental Microbiology, 78(9): 3136-3144. doi:10.1128/AEM.07800-11.
- ↑ Petersen, Jillian M. et al. (2011-08-11). "Hydrogen is an energy source for hydrothermal vent symbioses". Nature 476 (7359): 176–180. doi:10.1038/nature10325. PMID 21833083. Bibcode: 2011Natur.476..176P.

- ↑ Kleiner, Manuel; Wentrup, Cecilia; Lott, Christian; Teeling, Hanno; Wetzel, Silke; Young, Jacque; Chang, Yun-Juan; Shah, Manesh et al. (2012-05-08). "Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use". Proceedings of the National Academy of Sciences of the United States of America 109 (19): E1173–E1182. doi:10.1073/pnas.1121198109. PMID 22517752.
- ↑ Woyke, Tanja; Teeling, Hanno; Ivanova, Natalia N.; Huntemann, Marcel; Richter, Michael; Gloeckner, Frank Oliver; Boffelli, Dario; Anderson, Iain J. et al. (2006-10-26). "Symbiosis insights through metagenomic analysis of a microbial consortium". Nature 443 (7114): 950–955. doi:10.1038/nature05192. PMID 16980956. Bibcode: 2006Natur.443..950W. https://digital.library.unt.edu/ark:/67531/metadc879977/m2/1/high_res_d/914504.pdf.
- ↑ USGS scientists publish long-read microbiome sequences from temperate coral, providing community resource for probe and primer design, United States Geological Survey, 6 March 2019.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ b. Goldsmith, Dawn; a. Pratte, Zoe; a. Kellogg, Christina; e. Snader, Sara; h. Sharp, Koty (2019). "Stability of temperate coral Astrangia poculata microbiome is reflected across different sequencing methodologies". AIMS Microbiology 5 (1): 62–76. doi:10.3934/microbiol.2019.1.62. PMID 31384703.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ 174.0 174.1 Ruiz-González, C., Simó, R., Sommaruga, R. and Gasol, J.M. (2013) "Away from darkness: a review on the effects of solar radiation on heterotrophic bacterioplankton activity". Frontiers in microbiology, 4: 131. doi:10.3389/fmicb.2013.00131.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 3.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 3.0 International License.
- ↑ Walker, J. C. G. (1980). The Oxygen Cycle in the Natural Environment and the Biogeochemical Cycles. Berlin: Springer-Verlag.
- ↑ Longhurst, A., Sathyendranath, S., Platt, T., and Caverhill, C. (1995). An estimate of global primary production in the ocean from satellite radiometer data. J. Plankton Res. 17, 1245–1271.
- ↑ 177.0 177.1 177.2 177.3 177.4 Kurata, N., Vella, K., Hamilton, B., Shivji, M., Soloviev, A., Matt, S., Tartar, A. and Perrie, W. (2016) "Surfactant-associated bacteria in the near-surface layer of the ocean". Scientific Reports, 6(1): 1–8. doi:10.1038/srep19123. 50px Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Ẑutić, V., Ćosović, B., Marčenko, E., Bihari, N. and Kršinić, F. (1981) "Surfactant production by marine phytoplankton". Marine Chemistry, 10(6): 505–520. doi:10.1016/0304-4203(81)90004-9.
- ↑ 179.0 179.1 Siegel, David A.; Buesseler, Ken O.; Behrenfeld, Michael J.; Benitez-Nelson, Claudia R.; Boss, Emmanuel; Brzezinski, Mark A.; Burd, Adrian; Carlson, Craig A. et al. (2016). "Prediction of the Export and Fate of Global Ocean Net Primary Production: The EXPORTS Science Plan". Frontiers in Marine Science 3. doi:10.3389/fmars.2016.00022.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ "Diversity and distribution of Archaea in global estuarine ecosystems". The Science of the Total Environment 637–638: 349–358. October 2018. doi:10.1016/j.scitotenv.2018.05.016. PMID 29753224. Bibcode: 2018ScTEn.637..349L.
- ↑ "Nitrate reduction and the nitrogen cycle in archaea". Microbiology 150 (Pt 11): 3527–46. November 2004. doi:10.1099/mic.0.27303-0. PMID 15528644. http://mic.sgmjournals.org/cgi/content/full/150/11/3527?view=long&pmid=15528644.
- ↑ "Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon". Science 314 (5806): 1783–86. December 2006. doi:10.1126/science.1134772. PMID 17170307. Bibcode: 2006Sci...314.1783M.
- ↑ "New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation". The ISME Journal 1 (1): 19–27. May 2007. doi:10.1038/ismej.2007.8. PMID 18043610.
- ↑ "Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids". Environmental Microbiology 9 (4): 1001–16. April 2007. doi:10.1111/j.1462-2920.2006.01227.x. PMID 17359272.
- ↑ "Microbial communities in acid mine drainage". FEMS Microbiology Ecology 44 (2): 139–52. May 2003. doi:10.1016/S0168-6496(03)00028-X. PMID 19719632.
- ↑ "Playing scales in the methane cycle: from microbial ecology to the globe". Proceedings of the National Academy of Sciences of the United States of America 101 (34): 12400–01. August 2004. doi:10.1073/pnas.0405075101. PMID 15314221. Bibcode: 2004PNAS..10112400S.
Bibliography
- Dalrymple, G. Brent (2001). "The age of the Earth in the twentieth century: a problem (mostly) solved". Special Publications, Geological Society of London 190 (1): 205–221. doi:10.1144/GSL.SP.2001.190.01.14. Bibcode: 2001GSLSP.190..205D.
- Raven, Peter Hamilton; Johnson, George Brooks (2002). Biology. McGraw-Hill Education. ISBN 978-0-07-112261-0. https://archive.org/details/biologyrave00rave.
- Shors, Teri (2017). Understanding viruses. Jones and Bartlett Publishers. ISBN 978-1284025927.
 |
 KSF
KSF






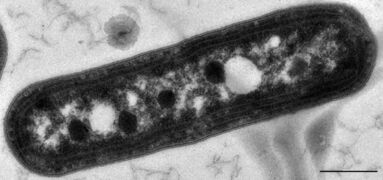
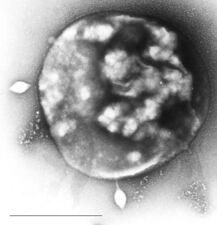
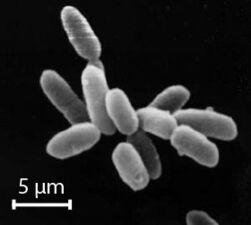



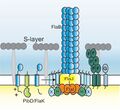
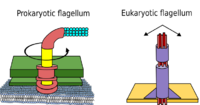
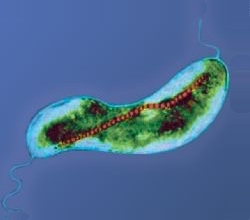



![Hesiocaeca methanicola lives at great depths on methane ice and appear to survive in symbiosis with bacteria which metabolize the clathrate.[166]](https://handwiki.org/wiki/images/thumb/b/bb/Hesiocaeca_methanicola_noaa.jpg/361px-Hesiocaeca_methanicola_noaa.jpg)