Murine coronavirus
Topic: Biology
 From HandWiki - Reading time: 12 min
From HandWiki - Reading time: 12 min
| Murine coronavirus | |
|---|---|
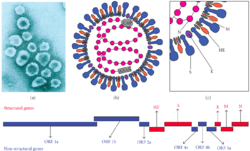
| |
| Murine coronavirus (MHV) virion electron micrograph, schematic structure, and genome | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Embecovirus |
| Species: | Murine coronavirus
|
| Strains | |
| |
Murine coronavirus (M-CoV) is a virus in the genus Betacoronavirus that infects mice.[3] Belonging to the subgenus Embecovirus,[4] murine coronavirus strains are enterotropic or polytropic. Enterotropic strains include mouse hepatitis virus (MHV) strains D, Y, RI, and DVIM, whereas polytropic strains, such as JHM and A59, primarily cause hepatitis, enteritis, and encephalitis.[5] Murine coronavirus is an important pathogen in the laboratory mouse and the laboratory rat. It is the most studied coronavirus in animals other than humans, and has been used as an animal disease model for many virological and clinical studies.[6]
Types
Murine hepatitis virus
Murine coronavirus was first discovered in 1949. The researchers isolated the virus from the brain, spinal cord, liver, lung, spleen, and kidney of a rat with symptoms of encephalitis and severe myelin injury, and gave it the strain name mouse hepatitis virus (MHV)-JHM.[7] MHV is now the most studied coronavirus in animals other than humans,[8] acting as a model organism for coronaviruses.[9]
There are more than 25 different strains of murine coronavirus. Transmitted by the fecal–oral or respiratory route, these viruses infect the livers of mice and have been used as an animal disease model for hepatitis.[10] Transmitted in fecal matter, the strains MHV-D, MHV-DVIM, MHV-Y and MHV-RI mainly infect the digestive tract, sometimes infecting the spleen, liver and lymphatic tissue.[8] MHV-1, MHV-2, MHV-3, MHV-A59, MHV-S, MHV-JHM and other virus strains replicate in the respiratory tract and then spread to other organs such as the liver, lungs and brain. [citation needed] MHV-JHM mainly infects the central nervous system and has been widely studied since 1949. In rats, these nerve-infecting hepatitis viruses can cause acute or chronic neurological symptoms[11] and stimulate the immunity of mice upon infection.[citation needed] Infection leads to demyelination, serving as an animal disease model of multiple sclerosis.[12] MHV-2, MHV-3 and MHV-A59 can also infect the liver; the first two of these are more virulent. MHV-3 is the main virus strain used to study hepatitis; MHV-1 mainly infects the lungs.[13]
Murine hepatitis virus is highly infectious and is one of the most common pathogens in laboratory mice. The symptoms of infection vary according to the type, path of infection, genotype and age of mouse. MHV-1, MHV-S and MHV-Y are weak viral strains; MHV-2, MHV-3, MHV-A5 9 and MHV-JHM are more virulent, being relatively mild in adult mice but having a high mortality in newborns.[8] Infection, even if it does not cause obvious symptoms, may affect the immune system of laboratory subjects and cause errors in the interpretation of experimental results.[14] For example, the virus can replicate in macrophages and affect their function, as well as in the spleen, where infection stimulates natural killer cells and affects T cell and B cells. There is no vaccine to prevent and treat hepatitis virus infection in mice, mainly because of the high mutation rate and the variety of virus strains, as well as concerns that vaccination may itself interfere with the interpretation of experimental research results, but this virus can be used as an experimental model for the development of other coronavirus vaccines.[8]
In 1991, Michael M. C. Lai's laboratory completed the whole genome sequencing of the murine hepatitis virus. With a total length of 31,000 nucleotides, it was the largest RNA virus genome known at that time.[15] In 2002, American virologist Ralph S. Baric developed a reverse genetic system for mouse hepatitis virus in which a complete MHV cDNA was assembled from smaller fragments.[16]
Fancy rat coronavirus
In fancy rats, the rat coronavirus (RCoV or RCV) consists mainly of two virus strains, sialodacryoadenitis virus (SDAV) and Parker's RCoV (RCoV-P), both of which cause respiratory tract infections, with the former also affecting the eyes, Harderian gland, and salivary glands. In the past, it was believed that the symptoms caused by the two infections were different, but in recent years, it has been argued that the symptoms of both include eye and nasal discharge, large salivary gland enlargement, sialadenitis, photosensitivity, keratitis, shortness of breath, and pneumonia, among others.[17][18][19] There is little to no obvious difference,[20] and it has been suggested that Parker's rat coronavirus is only one type of rat salivary adenovirus.[19] It is highly infectious. Generally, the symptoms in young rats are more serious, and some individuals suffer permanent eye damage.[19]
Others
In 1982, researchers found a coronavirus in the brains of mice after isolation of puffinosis coronavirus (PCoV), which causes skin and eye disease in Manx shearwaters. The virus found was very similar to rat hepatitis virus, but due to the use of laboratory mice in the isolation process, the possibility cannot be excluded that it was derived from a mouse and not from the birds.[21] Subsequent studies have shown that the virus has hemagglutinin esterase (HE).[22] If the coronavirus did indeed originate from shearwater, it is one of few bird coronaviruses that is not a gammacoronavirus or deltacoronavirus.[23] In 2009, the International Committee on Taxonomy of Viruses (ICTV) classified this bird coronavirus as belonging to the murine coronavirus clade.[2]
From 2011 to 2013, researchers collected mouse samples at several locations in Zhejiang, China, and discovered three new virus strains in Longquan lesser ricefield rat, collectively described in 2015 as Longquan Rl rat coronavirus (LRLV).[24]
Genome
Rat coronavirus is a positive-stranded single-strand RNA virus with an outer membrane. It has a genome size of about 31,000 nucleotides. In addition to the four structural proteins of coronaviruses — spike protein (S), membrane protein (M), envelope protein (E) and nucleocapsid protein (N) — some mouse coronavirus surfaces also have hemagglutinin esterase (HE). HE can bind to sialic acid on the surface of the host cell and promote viral infection, and has acetyl esterase activity, which can degrade receptors to release the bound virus.[11] The virus also has four auxiliary proteins — 2a, 4, 5a and I (or N2) (known as NS2, 15k, 12.6k and 7b[17] in rat salivary adenophritis virus or as 2a, 5a, 5b and N2 in Longquan Luosai mouse coronavirus[24]). These auxiliary proteins may counter the host's immune response. The auxiliary protein NS2 (encoded by the 2a gene) has 2′,5′-phosphodiesterase activity; it can degrade 2′,5′-oligoadenylate in the cell and avoid its activation. Ribonuclease L in cells activates the defense mechanism for degrading viral RNA[25] and auxiliary protein 5a inhibits host interferon.[26] The types of auxiliary proteins in different virus strains may differ. For example, MHV-S lacks auxiliary protein 5a, so it is less resistant to interferon.[26] All four auxiliary proteins are dispensable for viral replication.[27][28] The E protein is divided into the E1 and E2 glycoproteins, which are believed to serve different purposes.[29] The genome is ordered 1ab-2a-HE-S-4-5a-E-M-N-I, where 5a and 5b proteins are encoded by the same mRNA[27] and the open reading frame of I is located within the open reading frame of capsid protein N.[30]
Infection
When coronavirus infects the host cell, its spike protein (S) binds to the receptor on the surface of the host cell, which enables the virus to enter the cell. The spike protein is cut by the host's protease at all stages of the formation, transportation and infection of the new cell. The domain that helps the external membrane of the virus fuse with the cell membrane is exposed to facilitate infection. The host cell receptor used by rat coronavirus is generally CEACAM1 (mCEACAM1). The type of infected tissue and the time at which the spike protein is cut vary according to the virus strain. Among them is S1 in the spike protein of MHV-A59. The cleavage site of S2 is cut by proteases such as furin in the host cell when the virus is produced and assembled, and when the virus infects a new cell, further cleavage in the lysosomal pathway is also required for successful infection.[31] The ocyrosin of MHV-2 does not have the S1/S2 cleavage site and is not cut during the assembly process. Its infection depends on cleavage of the spike protein by endosomal enzymes.[32] MHV-JHM (especially the more virulent JHM.SD and JHM-cl2), which infects nerve tissue, may not require surface exposure[clarification needed]. The body can infect the cell[clarification needed], that is, it can achieve membrane fusion without binding to the cell receptor, so it can infect structures in the nervous system with little expression of mCEACAM1,[33][34] and its infection may mainly depend on the cutting of its spike protein by the cell surface protease.[35]
When rat hepatitis viruses of different strains infects cells at the same time, template switching can occur while genetic replication is carried out, resulting in gene recombination, which may be important for the evolution of viral diversity.[36][37]
Classification and evolution
This Classification and evolution section needs attention from an expert in virology. The specific problem is: proposed evolutionary sequence of events in 2nd paragraph of this section unclear. (January 2022) |
Murine coronavirus is believed to be most closely related to human coronavirus HKU1.[38] These two species, along with Betacoronavirus 1, rabbit coronavirus HKU14, and China Rattus coronavirus HKU24, form subgenus Embecovirus[39] within genus Betacoronavirus, according to the classification from the International Committee on Taxonomy of Viruses. This subgenus is distinguished by the presence of a gene encoding hemaglutinin esterase (HE),[38][40] although in many laboratory murine hepatitis virus strains (such as MHV-A59 and MHV-1), this gene has been lost to mutation and persists only as a pseudogene. HE is dispensable for rat hepatitis virus infection and replication,[41] and indeed, hepatitis strains lacking HE appear to have a competitive advantage in vitro.[42]
The N-terminal domain (NTD) of the spike protein of coronavirus is similar to galectin in animal cells.[43] Therefore, it has been suggested that this domain was originally derived from a host animal cell. The cell acquires the gene for a lectin, which can bind to the sugar on the surface of the host cell as an infected cell. Subsequently, the virus in this clade of coronaviruses acquires HE to help the virus get rid of infected cells, but later the NTD of the mouse coronavirus evolved into a new structure that can be associated with the protein receptor mCEACAM1. Combination greatly increases the binding ability of viruses and murine cells. Because it is no longer necessary to bind to sugars, it gradually loses the lectin function, and further loses the HE. In contrast, bovine coronavirus, human coronavirus OC43, and others are still sugar receptors, so the spike NTD retains the function of glutin.[44]
Alphacoronaviruses and betacoronaviruses may all originate from bat viruses, but the subgenus Embecovirus contains many viruses infecting rats (in addition to mouse coronavirus, there are also the Lucheng Rn rat coronavirus, China Rattus coronavirus HKU24 and Myodes coronavirus 2JL14, with a large number of related virus strains[45] found since 2015), and no bat virus has been found. Some scholars suggest that the common ancestor of this clade may be a mouse virus, which was then transmitted by rats to humans and cattle.[45][46]
RNA–RNA recombination
Genetic recombination can occur when at least two RNA viral genomes are present in the same infected host cell. RNA–RNA recombination between different strains of the murine coronavirus was found to occur at a high frequency both in tissue culture[47] and in the mouse central nervous system.[36] These findings suggest that RNA–RNA recombination may play a significant role in the natural evolution and neuropathogenesis of coronaviruses.[36] The mechanism of recombination appears to involve template switching during viral genome replication, a process referred to as copy choice recombination.[36]
Strains
Sialodacryoadenitis virus[48] is a highly infectious coronavirus of laboratory rats that can be transmitted between individuals by direct contact and indirectly by aerosol. Acute infections have high morbidity and tropism for the salivary, lachrymal and Harderian glands.
Rabbit enteric coronavirus causes acute gastrointestinal disease and diarrhea in young European rabbits.[49] Mortality rates are high.[50]
Research
Infection of mice with mouse hepatitis virus has been used as a model system to examine ivermectin as a treatment for coronaviruses.[51]
References
- ↑ ICTV 2nd Report Fenner, F. (1976). Classification and nomenclature of viruses. Second report of the International Committee on Taxonomy of Viruses. Intervirology 7: 1–115. https://ictv.global/ictv/proposals/ICTV%202nd%20Report.pdf
- ↑ 2.0 2.1 2.2 2.3 "Revision of the family Coronaviridae" (in en). 2009. p. 36. https://ictv.global/ictv/proposals/2008.085-122V.v4.Coronaviridae.pdf. "Species Murine hepatitis virus; Puffinosis coronavirus; Rat coronavirus (these are to be united in a new species Murine coronavirus in a new genus Betacoronavirus)"
- ↑ "Biology and Diseases of Rats". Laboratory Animal Medicine. Elsevier. 2015. pp. 151–207. doi:10.1016/b978-0-12-409527-4.00004-3. ISBN 978-0-12-409527-4.
- ↑ "Diversity of Dromedary Camel Coronavirus HKU23 in African Camels Revealed Multiple Recombination Events among Closely Related Betacoronaviruses of the Subgenus Embecovirus". Journal of Virology 93 (23). December 2019. doi:10.1128/JVI.01236-19. PMID 31534035.
- ↑ "Role of cytotoxic T lymphocytes and interferon-γ in coronavirus infection: Lessons from murine coronavirus infections in mice". The Journal of Veterinary Medical Science 82 (10): 1410–1414. October 2020. doi:10.1292/jvms.20-0313. PMID 32759577.
- ↑ "Of Mice and Men: The Coronavirus MHV and Mouse Models as a Translational Approach to Understand SARS-CoV-2". Viruses 12 (8): 880. August 2020. doi:10.3390/v12080880. PMID 32806708.
- ↑ "A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin". The Journal of Experimental Medicine 90 (3): 181–210. September 1949. doi:10.1084/jem.90.3.181. PMID 18137294.
- ↑ 8.0 8.1 8.2 8.3 "Of Mice and Men: The Coronavirus MHV and Mouse Models as a Translational Approach to Understand SARS-CoV-2". Viruses 12 (8): 880. August 2020. doi:10.3390/v12080880. PMID 32806708.
- ↑ "Forty years with coronaviruses". The Journal of Experimental Medicine 217 (5). May 2020. doi:10.1084/jem.20200537. PMID 32232339.
- ↑ "Acute and chronic changes in the microcirculation of the liver in inbred strains of mice following infection with mouse hepatitis virus type 3". Hepatology 5 (4): 649–60. 1985. doi:10.1002/hep.1840050422. PMID 2991107.
- ↑ 11.0 11.1 "Pathogenesis of murine coronavirus in the central nervous system". Journal of Neuroimmune Pharmacology 5 (3): 336–54. September 2010. doi:10.1007/s11481-010-9202-2. PMID 20369302.
- ↑ "Cell replacement therapies to promote remyelination in a viral model of demyelination". Journal of Neuroimmunology 224 (1–2): 101–7. July 2010. doi:10.1016/j.jneuroim.2010.05.013. PMID 20627412.
- ↑ "Coronavirus pathogenesis". Advances in Virus Research 81: 85–164. 2011. doi:10.1016/B978-0-12-385885-6.00009-2. ISBN 9780123858856. PMID 22094080.
- ↑ "Mouse Hepatitis Virus (MHV)". Division of Animal Resources,University of Illinois, Urbana. https://research.illinois.edu/files/upload/mhv.pdf.
- ↑ "The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase". Virology 180 (2): 567–82. February 1991. doi:10.1016/0042-6822(91)90071-I. PMID 1846489.
- ↑ "Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59". Journal of Virology 76 (21): 11065–78. November 2002. doi:10.1128/jvi.76.21.11065-11078.2002. PMID 12368349.
- ↑ 17.0 17.1 "Primary structure of the sialodacryoadenitis virus genome: sequence of the structural-protein region and its application for differential diagnosis". Clinical and Diagnostic Laboratory Immunology 7 (4): 568–73. July 2000. doi:10.1128/CDLI.7.4.568-573.2000. PMID 10882653.
- ↑ "Rat coronaviruses infect rat alveolar type I epithelial cells and induce expression of CXC chemokines". Virology 369 (2): 288–98. December 2007. doi:10.1016/j.virol.2007.07.030. PMID 17804032.
- ↑ 19.0 19.1 19.2 "Coronaviridae". Fenner's Veterinary Virology. Elsevier. 2017. pp. 435–461. doi:10.1016/b978-0-12-800946-8.00024-6. ISBN 978-0-12-800946-8.
- ↑ "Biology and Diseases of Rats". Laboratory Animal Medicine. Elsevier. 2002. pp. 121–165. doi:10.1016/b978-012263951-7/50007-7. ISBN 978-0-12-263951-7.
- ↑ "Isolation of a coronavirus during studies on puffinosis, a disease of the Manx shearwater (Puffinus puffinus)". Archives of Virology 73 (1): 1–13. 1982. doi:10.1007/BF01341722. PMID 7125912.
- ↑ "Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus". Journal of Virology 73 (5): 3737–43. May 1999. doi:10.1128/JVI.73.5.3737-3743.1999. PMID 10196267.
- ↑ "Coronaviruses in poultry and other birds". Avian Pathology 34 (6): 439–48. December 2005. doi:10.1080/03079450500367682. PMID 16537157.
- ↑ 24.0 24.1 "Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China". Virology 474: 19–27. January 2015. doi:10.1016/j.virol.2014.10.017. PMID 25463600.
- ↑ "Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology". Cell Host & Microbe 11 (6): 607–16. June 2012. doi:10.1016/j.chom.2012.04.011. PMID 22704621.
- ↑ 26.0 26.1 "Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus". Journal of Virology 84 (16): 8262–74. August 2010. doi:10.1128/JVI.00385-10. PMID 20519394.
- ↑ 27.0 27.1 "Mouse hepatitis virus S RNA sequence reveals that nonstructural proteins ns4 and ns5a are not essential for murine coronavirus replication". Journal of Virology 65 (10): 5605–8. October 1991. doi:10.1128/JVI.65.10.5605-5608.1991. PMID 1654456.
- ↑ "The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication". Journal of Virology 71 (2): 996–1003. February 1997. doi:10.1128/JVI.71.2.996-1003.1997. PMID 8995618.
- ↑ Fleming, J O; Trousdale, M D; el-Zaatari, F A; Stohlman, S A; Weiner, L P (June 1986). "Pathogenicity of antigenic variants of murine coronavirus JHM selected with monoclonal antibodies". Journal of Virology 58 (3): 869–875. doi:10.1128/jvi.58.3.869-875.1986. PMID 3701929.
- ↑ "Coronaviruses: An Overview of Their Replication and Pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. New York, NY: Springer New York. 2015. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2437-0.
- ↑ "Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner". PLOS Pathogens 10 (11): e1004502. November 2014. doi:10.1371/journal.ppat.1004502. PMID 25375324.
- ↑ "Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry". Journal of Virology 80 (12): 5768–76. June 2006. doi:10.1128/JVI.00442-06. PMID 16731916.
- ↑ "Structure, Function, and Evolution of Coronavirus Spike Proteins". Annual Review of Virology 3 (1): 237–261. September 2016. doi:10.1146/annurev-virology-110615-042301. PMID 27578435.
- ↑ "The spike glycoprotein of murine coronavirus MHV-JHM mediates receptor-independent infection and spread in the central nervous systems of Ceacam1a−/− Mice". Journal of Virology 82 (2): 755–63. January 2008. doi:10.1128/JVI.01851-07. PMID 18003729.
- ↑ "Neurovirulent Murine Coronavirus JHM.SD Uses Cellular Zinc Metalloproteases for Virus Entry and Cell–Cell Fusion". Journal of Virology 91 (8). April 2017. doi:10.1128/JVI.01564-16. PMID 28148786.
- ↑ 36.0 36.1 36.2 36.3 Keck JG, Matsushima GK, Makino S, Fleming JO, Vannier DM, Stohlman SA, Lai MM. In vivo RNA–RNA recombination of coronavirus in mouse brain. J Virol. 1988 May;62(5):1810–3. PMID 2833625
- ↑ "Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses". Trends in Microbiology 24 (6): 490–502. June 2016. doi:10.1016/j.tim.2016.03.003. PMID 27012512.
- ↑ 38.0 38.1 "Coronavirus genomics and bioinformatics analysis". Viruses 2 (8): 1804–20. August 2010. doi:10.3390/v2081803. PMID 21994708. "In all members of Betacoronavirus subgroup A, a haemagglutinin esterase (HE) gene, which encodes a glycoprotein with neuraminate O-acetyl-esterase activity and the active site FGDS, is present downstream to ORF1ab and upstream to S gene (Figure 1).".
- ↑ "Virus Taxonomy: 2018 Release". International Committee on Taxonomy of Viruses (ICTV). October 2018. https://ictv.global/taxonomy.
- ↑ "Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A". Journal of Virology 89 (6): 3076–92. March 2015. doi:10.1128/JVI.02420-14. PMID 25552712.
- ↑ "Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses". Virology 183 (2): 647–57. August 1991. doi:10.1016/0042-6822(91)90994-M. PMID 1649505.
- ↑ "Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro". Journal of Virology 79 (24): 15054–63. December 2005. doi:10.1128/JVI.79.24.15054-15063.2005. PMID 16306576.
- ↑ Caetano-Anollés, Kelsey; Hernandez, Nicolas; Mughal, Fizza; Tomaszewski, Tre; Caetano-Anollés, Gustavo (2022). "The seasonal behaviour of COVID-19 and its galectin-like culprit of the viral spike". Covid-19: Biomedical Perspectives. Methods in Microbiology. 50. pp. 27–81. doi:10.1016/bs.mim.2021.10.002. ISBN 9780323850612. https://doi.org/10.1016/bs.mim.2021.10.002.
- ↑ "Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor". Proceedings of the National Academy of Sciences of the United States of America 108 (26): 10696–701. June 2011. doi:10.1073/pnas.1104306108. PMID 21670291. Bibcode: 2011PNAS..10810696P.
- ↑ 45.0 45.1 "Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases". Microbiome 6 (1): 178. October 2018. doi:10.1186/s40168-018-0554-9. PMID 30285857.
- ↑ "Molecular Evolution of Human Coronavirus Genomes". Trends in Microbiology 25 (1): 35–48. January 2017. doi:10.1016/j.tim.2016.09.001. PMID 27743750. "Specifically, all HCoVs are thought to have a bat origin, with the exception of lineage A beta-CoVs, which may have reservoirs in rodents [2].".
- ↑ Makino S, Keck JG, Stohlman SA, Lai MM. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986 Mar;57(3):729–37. PMID 3005623
- ↑ "Taxonomy browser (Embecovirus)". https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=2509481&lvl=3&lin=f&keep=1&srchmode=1&unlock.
- ↑ "Chapter 24 – Coronaviridae" (in en). Fenner's Veterinary Virology (Fifth ed.). Academic Press. 2017. pp. 435–461. doi:10.1016/B978-0-12-800946-8.00024-6. ISBN 978-0-12-800946-8.
- ↑ "Enteric Coronavirus". http://dora.missouri.edu/rabbits/enteric-coronavirus/.
- ↑ Arévalo, A. P.; Pagotto, R.; Pórfido, J. L.; Daghero, H.; Segovia, M.; Yamasaki, K.; Varela, B.; Hill, M. et al. (December 2021). "Ivermectin reduces in vivo coronavirus infection in a mouse experimental model". Scientific Reports 11 (1): 7132. doi:10.1038/s41598-021-86679-0. PMID 33785846. Bibcode: 2021NatSR..11.7132A.
Wikidata ☰ Q18907882 entry
 |
 KSF
KSF