Systems Biology Graphical Notation
Topic: Biology
 From HandWiki - Reading time: 6 min
From HandWiki - Reading time: 6 min

The Systems Biology Graphical Notation (SBGN) is a standard graphical representation intended to foster the efficient storage, exchange and reuse of information about signaling pathways, metabolic networks, and gene regulatory networks amongst communities of biochemists, biologists, and theoreticians. The system was created over several years by a community of biochemists, modelers and computer scientists.[1]
SBGN is made up of three orthogonal languages for representing different views of biological systems: Process Descriptions, Entity Relationships and Activity Flows. Each language defines a comprehensive set of symbols with precise semantics, together with detailed syntactic rules regarding the construction and interpretation of maps. Using these three notations, a life scientist can represent in an unambiguous way networks of interactions (for example biochemical interactions). These notations make use of an idea and symbols similar to that used by electrical and other engineers and known as the block diagram. The simplicity of SBGN syntax and semantics makes SBGN maps suitable for use at the high school level.[citation needed]
Some software support for SBGN is already available, mostly for the Process Description language.[2] SBGN visualizations can be exchanged with the XML-based file format SBGN-ML.[3]
SBGN Process Description language
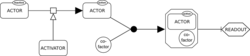
The SBGN Process Description (PD) language shows the temporal courses of biochemical interactions in a network. It can be used to show all the molecular interactions taking place in a network of biochemical entities, with the same entity appearing multiple times in the same diagram.[4]
SBGN Entity Relationship language
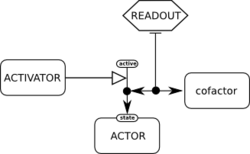
The SBGN Entity Relationship (ER) language allows to see all the relationships in which a given entity participates, regardless of the temporal aspects. Relationships can be seen as rules describing the influences of entities nodes on other relationships.[5]
SBGN Activity Flow language

The SBGN Activity Flow (AF) language depicts the flow of information between biochemical entities in a network. It omits information about the state transitions of entities and is particularly convenient for representing the effects of perturbations, whether genetic or environmental in nature.[6]
History
Work on defining a set of symbols to describe interactions and relationships of molecules was pioneered by Kurt Kohn at the National Cancer Institute with his Molecular Interaction Maps (MIM).[7] The development of SBGN was initiated by Hiroaki Kitano, supported by a funding from the Japanese New Energy and Industrial Technology Development Organization. The meeting that initiated development of the Systems Biology Graphical Notation took place on February 11–12, 2006, at the National Institute of Advanced Industrial Science and Technology (AIST), in Tokyo, Japan.
The first specification of SBGN Process Description language – then called Process Diagrams – was released on August 23, 2008 (Level 1 Version 1).[8] Corrections of the document were released on September 1, 2009 (Level 1 Version 1.1),[9] October 3, 2010 (Level 1 Version 1.2)[10] and February 14, 2011 (Level 1 Version 1.3).[4]
The first specification of SBGN Entity relationship language was released on September 1, 2009 (Level 1 Version 1).[11] Corrections of the document were released on October 6, 2010 (Level 1 Version 1.1)[12] and April 14, 2011 (Level 1 Version 1.2).[5]
The first specification of SBGN Activity Flow language was released on September 1, 2009.[6]
Editors
SBGN editors work in developing coherent specification documents. Below is a list of former SBGN editors and dates active:[13]
- Ugur Dogrusoz (Jan 2022 - Dec 2024)
- Adrien Rougn (Jan 2022 - Dec 2024)
- Alexander Mazein (Jan 2021 - Dec 2023)
- Irina Balaur (Jan 2021 - Dec 2023)
- Emek Demir (Jan 2020 - Dec 2022)
- Andreas Dräger (Jan 2018 - Dec 2021)
- Falk Schreiber (Sep 2008 - Dec 2010, Jan 2012 - Dec 2014)
- Anatoly Sorokin (Sep 2008 - Dec 2009, Jan 2012 - Dec 2014)
- Stuart Moodie (Sep 2008 - Dec 2011, Sep 2012 - Dec 2015)
- Nicolas Le Novère (Sep 2008 - Dec 2012, Dec 2015 - Dec 2016)
- Emek Demir (Jan 2010 - Dec 2012)
- Alice Villéger (Jan 2011 - Dec 2013)
- Huaiyu Mi (Jan 2009 - Dec 2011, Dec 2013 - Dec 2016)
- Tobias Czauderna (Jan 2012 - Dec 2015, Dec 2016 - Dec 2017)
- Robin Haw (Dec 2014 - Dec 2017)
- Augustin Luna (Dec 2014 - Dec 2017)
- Alexander Mazein (Dec 2015 - Dec 2018)
- Ugur Dogrusoz (Dec 2016 - Dec 2019)
- Vasundra Touré (Jan 2018 - Feb 2021)
- Adrien Rougny (Jan 2018 - Feb 2021)
External links
References
- ↑ "The Systems Biology Graphical Notation.". Nat Biotechnol 27 (8): 735–41. 2009. doi:10.1038/nbt.1558. PMID 19668183.
- ↑ "Community#SBGN_Software". sbgn.org. Archived from the original on 2012-05-07. https://web.archive.org/web/20120507060310/http://www.sbgn.org/Community#SBGN_Software. Retrieved 2009-11-30.
- ↑ "Systems Biology Graphical Notation" (in en-US). https://sbgn.github.io/.
- ↑ 4.0 4.1 Moodie SL, Le Novère N, Demir E, Mi H & Villéger A. Systems Biology Graphical Notation: Process Description language Level 1. Nature Precedings (2011) doi:10.1038/npre.2011.3721.4
- ↑ 5.0 5.1 Le Novère N, Demir E, Mi H, Moodie S & Villéger A. Systems Biology Graphical Notation: Entity Relationship language Level 1. Nature Precedings (2010) doi:10.1038/npre.2011.5902.1
- ↑ 6.0 6.1 Mi H, Schreiber F, Le Novère N, Moodie S, Sorokin A. Systems Biology Graphical Notation: Activity Flow language Level 1. Nature Precedings (2009) doi:10.1038/npre.2009.3724.1
- ↑ Kohn KW, Aladjem MI, Weinstein JN, Pommier Y. Molecular interaction maps of bioregulatory networks: a general rubric for systems biology. Mol Biol Cell (2006) doi:10.1091/mbc.E05-09-0824
- ↑ Le Novère N, Moodie S, Sorokin A, Hucka M, Schreiber F, Demir E, Mi H, Matsuoka Y, Wegner K, Kitano H. Systems Biology Graphical Notation: Process Diagram Level 1. Nature Precedings (2008) doi:10101/npre.2008.2320.1
- ↑ Moodie S, Le Novère N, Sorokin A, Mi H, Schreiber F. Systems Biology Graphical Notation: Process Description language Level 1. Nature Precedings (2009) doi:10.1038/npre.2009.3721.1
- ↑ Moodie SL, Le Novère N, Demir E, Mi H, Schreiber F. Systems Biology Graphical Notation: Process Description language Level 1. Nature Precedings (2010) doi:10101/npre.2010.3721.2
- ↑ Le Novère N, Moodie S, Sorokin A, Schreiber F, Mi H. Systems Biology Graphical Notation: Entity Relationship language Level 1. Nature Precedings (2009) doi:10.1038/npre.2009.3719.1
- ↑ Le Novère N, Moodie S, Demir E, Schreiber F, Mi H. Systems Biology Graphical Notation: Entity Relationship language Level 1. Nature Precedings (2010) doi:10.1038/npre.2010.3719.2
- ↑ "About SBGN". 20 July 2021. https://sbgn.org/about#former-sbgn-editors.
 |
 KSF
KSF