Connectome
 From HandWiki - Reading time: 23 min
From HandWiki - Reading time: 23 min

A connectome (/kəˈnɛktoʊm/) is a comprehensive map of neural connections in the brain, and may be thought of as its "wiring diagram".[2] An organism's nervous system is made up of neurons which communicate through synapses. A connectome is constructed by tracing the neuron in a nervous system and mapping where neurons are connected through synapses.
The significance of the connectome stems from the realization that the structure and function of the human brain are intricately linked, through multiple levels and modes of brain connectivity. There are strong natural constraints on which neurons or neural populations can interact, or how strong or direct their interactions are. Indeed, the foundation of human cognition lies in the pattern of dynamic interactions shaped by the connectome.
Despite such complex and variable structure-function mappings, the connectome is an indispensable basis for the mechanistic interpretation of dynamic brain data, from single-cell recordings to functional neuroimaging.
Origin and usage of the term
In 2005, Dr. Olaf Sporns at Indiana University and Dr. Patric Hagmann at Lausanne University Hospital independently and simultaneously suggested the term "connectome" to refer to a map of the neural connections within the brain. This term was directly inspired by the ongoing effort to sequence the human genetic code—to build a genome.
"Connectomics" (Hagmann, 2005) has been defined as the science concerned with assembling and analyzing connectome data sets.[3]
In their 2005 paper, "The Human Connectome, a structural description of the human brain", Sporns et al. wrote:
To understand the functioning of a network, one must know its elements and their interconnections. The purpose of this article is to discuss research strategies aimed at a comprehensive structural description of the network of elements and connections forming the human brain. We propose to call this dataset the human "connectome," and we argue that it is fundamentally important in cognitive neuroscience and neuropsychology. The connectome will significantly increase our understanding of how functional brain states emerge from their underlying structural substrate, and will provide new mechanistic insights into how brain function is affected if this structural substrate is disrupted.[4]
In his 2005 Ph.D. thesis, From diffusion MRI to brain connectomics, Hagmann wrote:
It is clear that, like the genome, which is much more than just a juxtaposition of genes, the set of all neuronal connections in the brain is much more than the sum of their individual components. The genome is an entity it-self, as it is from the subtle gene interaction that [life] emerges. In a similar manner, one could consider the brain connectome, set of all neuronal connections, as one single entity, thus emphasizing the fact that the huge brain neuronal communication capacity and computational power critically relies on this subtle and incredibly complex connectivity architecture.[3]
The term "connectome" was more recently popularized by Sebastian Seung's I am my Connectome speech given at the 2010 TED conference, which discusses the high-level goals of mapping the human connectome, as well as ongoing efforts to build a three-dimensional neural map of brain tissue at the microscale.[5] In 2012, Seung published the book Connectome: How the Brain's Wiring Makes Us Who We Are.
Methods
Brain networks can be defined at different levels of scale, corresponding to levels of spatial resolution in brain imaging (Kötter, 2007, Sporns, 2010).[6][7] These scales can be roughly categorized as macroscale, mesoscale and microscale. Ultimately, it may be possible to join connectomic maps obtained at different scales into a single hierarchical map of the neural organization of a given species that ranges from single neurons to populations of neurons to larger systems like cortical areas. Given the methodological uncertainties involved in inferring connectivity from the primary experimental data, and given that there are likely to be large differences in the connectomes of different individuals, any unified map will likely rely on probabilistic representations of connectivity data (Sporns et al., 2005).[4]
Macroscale
A connectome at the macroscale (millimeter resolution) attempts to capture large brain systems that can be parcellated into anatomically distinct modules (areas, parcels or nodes), each having a distinct pattern of connectivity. Connectomic databases at the mesoscale and macroscale may be significantly more compact than those at cellular resolution, but they require effective strategies for accurate anatomical or functional parcellation of the neural volume into network nodes (for complexities see, e.g., Wallace et al., 2004).[8]
Established methods of brain research, such as axonal tracing, provided early avenues for building connectome data sets. However, more recent advances in living subjects has been made by the use of non-invasive imaging technologies such as diffusion-weighted magnetic resonance imaging (DW-MRI) and functional magnetic resonance imaging (fMRI). The first, when combined with tractography allows reconstruction of the major fiber bundles in the brain. The second allows the researcher to capture the brain's network activity (either at rest or while performing directed tasks), enabling the identification of structurally and anatomically distinct areas of the brain that are functionally connected.
Notably, the goal of the Human Connectome Project, led by the WU-Minn consortium, is to build a structural and functional map of the healthy human brain at the macro scale, using a combination of multiple imaging technologies and resolutions.
Recent advances in connectivity mapping
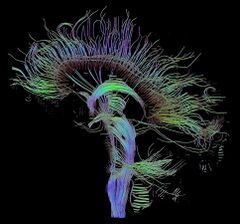
Throughout the 2000s, several investigators have attempted to map the large-scale structural architecture of the human cerebral cortex. One attempt exploited cross-correlations in cortical thickness or volume across individuals (He et al., 2007).[9] Such gray-matter thickness correlations have been postulated as indicators for the presence of structural connections. A drawback of the approach is that it provides highly indirect information about cortical connection patterns and requires data from large numbers of individuals to derive a single connection data set across a subject group. Other investigators have attempted to build whole-brain connection matrices from DW-MRI imaging data.
The Blue Brain Project is attempting to reconstruct the entire mouse connectome using a diamond knife sharpened to an atomic edge, and electron microscopy for imaging tissue slices.
Challenge for macroscale connectomics
The initial explorations in macroscale human connectomics were done using either equally sized regions or anatomical regions with unclear relationship to the underlying functional organization of the brain (e.g. gyral and sulcal-based regions). While much can be learned from these approaches, it is highly desirable to parcellate the brain into functionally distinct parcels: brain regions with distinct architectonics, connectivity, function, and/or topography (Felleman and Van Essen, 1991).[10] Accurate parcellation allows each node in the macroscale connectome to be more informative by associating it with a distinct connectivity pattern and functional profile. Parcellation of localized areas of cortex have been accomplished using diffusion tractography (Beckmann et al. 2009)[11] and functional connectivity (Nelson et al. 2010)[12] to non-invasively measure connectivity patterns and define cortical areas based on distinct connectivity patterns. Such analyses may best be done on a whole brain scale and by integrating non-invasive modalities. Accurate whole brain parcellation may lead to more accurate macroscale connectomes for the normal brain, which can then be compared to disease states.
Pathways through cerebral white matter can be charted by histological dissection and staining, by degeneration methods, and by axonal tracing. Axonal tracing methods form the primary basis for the systematic charting of long-distance pathways into extensive, species-specific anatomical connection matrices between gray matter regions. Landmark studies have included the areas and connections of the visual cortex of the macaque (Felleman and Van Essen, 1991)[10] and the thalamocortical system in the feline brain (Scannell et al., 1999).[13] The development of neuroinformatics databases for anatomical connectivity allow for continual updating and refinement of such anatomical connection maps. The online macaque cortex connectivity tool CoCoMac (Kötter, 2004)[14] and the temporal lobe connectome of the rat[15] are prominent examples of such a database.
Mesoscale
A "mesoscale" connectome corresponds to a spatial resolution of hundreds of micrometers. Rather than attempt to map each individual neuron, a connectome at the mesoscale would attempt to capture anatomically and/or functionally distinct neuronal populations, formed by local circuits (e.g. cortical columns) that link hundreds or thousands of individual neurons. This scale still presents a very ambitious technical challenge at this time and can only be probed on a small scale with invasive techniques or very high field magnetic resonance imaging (MRI) on a local scale.
Microscale
Mapping the connectome at the "microscale" (micrometer resolution) means building a complete map of the neural systems, neuron-by-neuron. The challenge of doing this becomes obvious: the number of neurons comprising the brain easily ranges into the billions in more complex organisms. The human cerebral cortex alone contains on the order of 1010 neurons linked by 1014 synaptic connections.[16] By comparison, the number of base-pairs in a human genome is 3×109. A few of the main challenges of building a human connectome at the microscale today include: data collection would take years given current technology, machine vision tools to annotate the data remain in their infancy, and are inadequate, and neither theory nor algorithms are readily available for the analysis of the resulting brain-graphs. To address the data collection issues, several groups are building high-throughput serial electron microscopes (Kasthuri et al., 2009; Bock et al. 2011). To address the machine-vision and image-processing issues, the Open Connectome Project[17] is alg-sourcing (algorithm outsourcing) this hurdle. Finally, statistical graph theory is an emerging discipline which is developing sophisticated pattern recognition and inference tools to parse these brain-graphs (Goldenberg et al., 2009).
Current non-invasive imaging techniques cannot capture the brain's activity on a neuron-by-neuron level. Mapping the connectome at the cellular level in vertebrates currently requires post-mortem (after death) microscopic analysis of limited portions of brain tissue. Non-optical techniques that rely on high-throughput DNA sequencing have been proposed recently by Anthony Zador (CSHL).[18]
Traditional histological circuit-mapping approaches rely on imaging and include light-microscopic techniques for cell staining, injection of labeling agents for tract tracing, or chemical brain preservation, staining and reconstruction of serially sectioned tissue blocks via electron microscopy (EM). Each of these classical approaches has specific drawbacks when it comes to deployment for connectomics. The staining of single cells, e.g. with the Golgi stain, to trace cellular processes and connectivity suffers from the limited resolution of light-microscopy as well as difficulties in capturing long-range projections. Tract tracing, often described as the "gold standard" of neuroanatomy for detecting long-range pathways across the brain, generally only allows the tracing of fairly large cell populations and single axonal pathways. EM reconstruction was successfully used for the compilation of the C. elegans connectome (White et al., 1986).[19] However, applications to larger tissue blocks of entire nervous systems have traditionally had difficulty with projections that span longer distances.
Recent advances in mapping neural connectivity at the cellular level offer significant new hope for overcoming the limitations of classical techniques and for compiling cellular connectome data sets (Livet et al., 2007; Lichtman et al., 2008).[20][21][22] Using Brainbow, a combinatorial color labeling method based on the stochastic expression of several fluorescent proteins, Jeff W. Lichtman and colleagues were able to mark individual neurons with one of over 100 distinct colors. The labeling of individual neurons with a distinguishable hue then allows the tracing and reconstruction of their cellular structure including long processes within a block of tissue.
In March 2011, the journal Nature published a pair of articles on micro-connectomes: Bock et al.[23] and Briggman et al.[24] In both articles, the authors first characterized the functional properties of a small subset of cells, and then manually traced a subset of the processes emanating from those cells to obtain a partial subgraph. In alignment with the principles of open science, the authors of Bock et al. (2011) have released their data for public access. The full resolution 12 terabyte dataset from Bock et al. is available at NeuroData.[17] Independently, important topologies of functional interactions among several hundred cells are also gradually going to be declared (Shimono and Beggs, 2014).[25] Scaling up ultrastructural circuit mapping to the whole mouse brain is currently underway (Mikula, 2012).[26] An alternative approach to mapping connectivity was recently proposed by Zador and colleagues (Zador et al., 2012).[18] Zador's technique, called BOINC (barcoding of individual neuronal connections) uses high-throughput DNA sequencing to map neural circuits. Briefly, the approach consists of labelling each neuron with a unique DNA barcode, transferring barcodes between synaptically coupled neurons (for example using Suid herpesvirus 1, SuHV1), and fusion of barcodes to represent a synaptic pair. This approach has the potential to be cheap, fast, and extremely high-throughput.
In 2016, the Intelligence Advanced Research Projects Activity of the United States government launched MICrONS, a five-year, multi-institute project to map one cubic millimeter of rodent visual cortex, as part of the BRAIN Initiative.[27][28] Though only a small volume of biological tissue, this project will yield one of the largest micro-scale connectomics datasets currently in existence.
Mapping functional connectivity
Using fMRI in the resting state and during tasks, functions of the connectome circuits are being studied.[29] Just as detailed road maps of the Earth's surface do not tell us much about the kind of vehicles that travel those roads or what cargo they are hauling, to understand how neural structures result in specific functional behavior such as consciousness, it is necessary to build theories that relate functions to anatomical connectivity.[30] However, the bond between structural and functional connectivity is not straightforward. Computational models of whole-brain network dynamics are valuable tools to investigate the role of the anatomical network in shaping functional connectivity.[31][32] In particular, computational models can be used to predict the dynamic effect of lesions in the connectome.[33][34]
As a network or graph
The connectome can be studied as a network by means of network science and graph theory. In case of a micro-scale connectome, the nodes of this network (or graph) are the neurons, and the edges correspond to the synapses between those neurons. For the macro-scale connectome, the nodes correspond to the ROIs (regions of interest), while the edges of the graph are derived from the axons interconnecting those areas. Thus connectomes are sometimes referred to as brain graphs, as they are indeed graphs in a mathematical sense which describe the connections in the brain (or, in a broader sense, the whole nervous system).
One group of researchers (Iturria-Medina et al., 2008)[35] has constructed connectome data sets using diffusion tensor imaging (DTI)[36][37] followed by the derivation of average connection probabilities between 70–90 cortical and basal brain gray matter areas. All networks were found to have small-world attributes and "broad-scale" degree distributions. An analysis of betweenness centrality in these networks demonstrated high centrality for the precuneus, the insula, the superior parietal and the superior frontal cortex. Another group (Gong et al. 2008)[38] has applied DTI to map a network of anatomical connections between 78 cortical regions. This study also identified several hub regions in the human brain, including the precuneus and the superior frontal gyrus.
Hagmann et al. (2007)[39] constructed a connection matrix from fiber densities measured between homogeneously distributed and equal-sized ROIs numbering between 500 and 4000. A quantitative analysis of connection matrices obtained for approximately 1,000 ROIs and approximately 50,000 fiber pathways from two subjects demonstrated an exponential (one-scale) degree distribution as well as robust small-world attributes for the network. The data sets were derived from diffusion spectrum imaging (DSI) (Wedeen, 2005),[40] a variant of diffusion-weighted imaging[41][42] that is sensitive to intra-voxel heterogeneities in diffusion directions caused by crossing fiber tracts and thus allows more accurate mapping of axonal trajectories than other diffusion imaging approaches (Wedeen, 2008).[43] The combination of whole-head DSI datasets acquired and processed according to the approach developed by Hagmann et al. (2007)[39] with the graph analysis tools conceived initially for animal tracing studies (Sporns, 2006; Sporns, 2007)[44][45] allow a detailed study of the network structure of human cortical connectivity (Hagmann et al., 2008).[46] The human brain network was characterized using a broad array of network analysis methods including core decomposition, modularity analysis, hub classification and centrality. Hagmann et al. presented evidence for the existence of a structural core of highly and mutually interconnected brain regions, located primarily in posterior medial and parietal cortex. The core comprises portions of the posterior cingulate cortex, the precuneus, the cuneus, the paracentral lobule, the isthmus of the cingulate, the banks of the superior temporal sulcus, and the inferior and superior parietal cortex, all located in both cerebral hemispheres.
A subfield of connectomics deals with the comparison of the brain graphs of multiple subjects. It is possible to build a consensus graph such the Budapest Reference Connectome by allowing only edges that are present in at least connectomes, for a selectable parameter. The Budapest Reference Connectome has led the researchers to the discovery of the Consensus Connectome Dynamics of the human brain graphs. The edges appeared in all of the brain graphs form a connected subgraph around the brainstem. By allowing gradually less frequent edges, this core subgraph grows continuously, as a shrub. The growth dynamics may reflect the individual brain development and provide an opportunity to direct some edges of the human consensus brain graph.[47]
Alternatively, local difference which are statistically significantly different among groups have attracted more attention as they highlight specific connections and therefore shed more light on specific brain traits or pathology. Hence, algorithms to find local difference between graph populations have also been introduced (e.g. to compare case versus control groups).[48] Those can be found by using either an adjusted t-test,[49] or a sparsity model,[48] with the aim of finding statistically significant connections which are different among those groups.
The possible causes of the difference between individual connectomes were also investigated. Indeed, it has been found that the macro-scale connectomes of women contain significantly more edges than those of men, and a larger portion of the edges in the connectomes of women run between the two hemispheres.[50][51][52] In addition, connectomes generally exhibit a small-world character, with overall cortical connectivity decreasing with age.[53] The aim of the as of 2015 ongoing HCP Lifespan Pilot Project is to identify connectome differences between 6 age groups (4–6, 8–9, 14–15, 25–35, 45–55, 65–75).
More recently, connectograms have been used to visualize full-brain data by placing cortical areas around a circle, organized by lobe.[54][55] Inner circles then depict cortical metrics on a color scale. White matter fiber connections in DTI data are then drawn between these cortical regions and weighted by fractional anisotropy and strength of the connection. Such graphs have even been used to analyze the damage done to the famous traumatic brain injury patient Phineas Gage.[56]
Statistical graph theory is an emerging discipline which is developing sophisticated pattern recognition and inference tools to parse these brain graphs (Goldenberg et al., 2009).
Recent research studied the brain as a signed network and indicated that hubness in positive and negative subnetworks increases the stability of the brain network. It highlighted the role of negative functional connections that are paid less attention to.[57]
Plasticity of the connectome
At the beginning of the connectome project, it was thought that the connections between neurons were unchangeable once established and that only individual synapses could be altered.[4] However, recent evidence suggests that connectivity is also subject to change, termed neuroplasticity. There are two ways that the brain can rewire: formation and removal of synapses in an established connection or formation or removal of entire connections between neurons.[58] Both mechanisms of rewiring are useful for learning completely novel tasks that may require entirely new connections between regions of the brain.[59] However, the ability of the brain to gain or lose entire connections poses an issue for mapping a universal species connectome. Although rewiring happens on different scales, from microscale to macroscale, each scale does not occur in isolation. For example, in the C. elegans connectome, the total number of synapses increases 5-fold from birth to adulthood, changing both local and global network properties.[60] Other developmental connectomes, such as the muscle connectome, retain some global network properties even though the number of synapses decreases by 10-fold in early postnatal life.[61]
Macroscale rewiring
Evidence for macroscale rewiring mostly comes from research on grey and white matter density, which could indicate new connections or changes in axon density. Direct evidence for this level of rewiring comes from primate studies, using viral tracing to map the formation of connections. Primates that were taught to use novel tools developed new connections between the interparietal cortex and higher visual areas of the brain.[62] Further viral tracing studies have provided evidence that macroscale rewiring occurs in adult animals during associative learning.[63] However, it is not likely that long-distance neural connections undergo extensive rewiring in adults. Small changes in an already established nerve tract are likely what is observed in macroscale rewiring.
Mesoscale rewiring
Rewiring at the mesoscale involves studying the presence or absence of entire connections between neurons.[59] Evidence for this level of rewiring comes from observations that local circuits form new connections as a result of experience-dependent plasticity in the visual cortex. Additionally, the number of local connections between pyramidal neurons in the primary somatosensory cortex increases following altered whisker sensory experience in rodents.[64]
Microscale rewiring
Microscale rewiring is the formation or removal of synaptic connections between two neurons and can be studied with longitudinal two-photon imaging. Dendritic spines on pyramidal neurons can be shown forming within days following sensory experience and learning.[65][66][67] Changes can even be seen within five hours on apical tufts of layer five pyramidal neurons in the primary motor cortex after a seed reaching task in primates.[67]
Datasets
Humans
The Human Connectome Project, sponsored by the National Institutes of Health (NIH), was created with the goal of mapping the 86 billion neurons (and their connections) in a human brain.[68]
Model organisms
Roundworm
The first (and so far only) fully reconstructed connectome belongs to the roundworm Caenorhabditis elegans.[69] The major effort began with the first electron micrographs published by White, Brenner et al., 1986.[19] Based on this seminal work, the first ever connectome (then called "neural circuitry database" by the authors) for C. elegans was published in book form with accompanying floppy disks by Achacoso and Yamamoto in 1992.[70][71] The very first paper on the computer representation of its connectome was presented and published three years earlier in 1989 by Achacoso at the Symposium on Computer Application in Medical Care (SCAMC).[72] The C. elegans connectome was later revised[73][74] and expanded to show changes during the animal's development.[60][75] Despite having an invariant cell lineage, the C. elegans connectome shows variability between individuals, both at the level of synapse and connection.[76][77]
Fruit fly
The fruit fly, Drosophila melanogaster, serves as an appealing model for exploring the structure and operation of nervous systems. Its central nervous system (CNS) is notably compact, housing approximately 200,000 neurons in adults, yet it exhibits reasonably stereotyped neural connections across individual flies.[78] Despite its small size, this CNS supports a broad spectrum of complex and well-studied behaviors. Obtaining an anatomical dataset of the fly's CNS could be a pivotal step, potentially offering insights into the nervous systems of other organisms.
A full electron microscopy (EM) connectome of the larval brain of D. melanogaster, including 3016 neurons and 548,000 synapses, was published in March 2023.[79][80][81] For adults, partial EM connectomes of the brain (~120,000 neurons, ~30,000,000 synapses)[82][83][84] or the ventral nerve cord (VNC, the fly's equivalent of the spinal cord, ~14,600 neurons)[85][86] are also available. A complete adult CNS connectome that includes both the brain and the VNC is currently under construction.
Mouse
Partial connectomes of a mouse retina[24] and mouse primary visual cortex[23] are available.
The first full connectome of a mammalian circuit was constructed in 2021. This construction included the development of all connections between the central nervous system and a single muscle from birth to adulthood.[61]
See also
References
- ↑ "The structural-functional connectome and the default mode network of the human brain". NeuroImage 102 Pt 1: 142–51. November 2014. doi:10.1016/j.neuroimage.2013.09.069. PMID 24099851.
- ↑ Mackenzie, Dana (6 March 2023). "How animals follow their nose" (in en). Knowable Magazine (Annual Reviews). doi:10.1146/knowable-030623-4. https://knowablemagazine.org/article/living-world/2023/how-animals-follow-their-nose. Retrieved 13 March 2023.
- ↑ 3.0 3.1 Hagmann, Patric (2005). From diffusion MRI to brain connectomics (Thesis). Lausanne: EPFL. doi:10.5075/epfl-thesis-3230. Retrieved 2014-01-16.
- ↑ 4.0 4.1 4.2 "The human connectome: A structural description of the human brain". PLOS Computational Biology 1 (4): e42. September 2005. doi:10.1371/journal.pcbi.0010042. PMID 16201007. Bibcode: 2005PLSCB...1...42S.

- ↑ Seung, Sebastian (September 2010). "Sebastian Seung: I am my connectome". TEDTalks. http://www.ted.com/talks/sebastian_seung.html.
- ↑ Kötter, Rolf (2007). "Anatomical Concepts of Brain Connectivity". Handbook of Brain Connectivity. Understanding Complex Systems. pp. 149–67. doi:10.1007/978-3-540-71512-2_5. ISBN 978-3-540-71462-0.
- ↑ Sporns, Olaf (2011). Networks of the Brain. Cambridge, Mass.: MIT Press. ISBN 978-0-262-01469-4.
- ↑ "A revised view of sensory cortical parcellation". Proceedings of the National Academy of Sciences of the United States of America 101 (7): 2167–72. February 2004. doi:10.1073/pnas.0305697101. PMID 14766982. Bibcode: 2004PNAS..101.2167W.
- ↑ "Small-world anatomical networks in the human brain revealed by cortical thickness from MRI". Cerebral Cortex 17 (10): 2407–19. October 2007. doi:10.1093/cercor/bhl149. PMID 17204824.
- ↑ 10.0 10.1 "Distributed hierarchical processing in the primate cerebral cortex". Cerebral Cortex 1 (1): 1–47. 1991. doi:10.1093/cercor/1.1.1-a. PMID 1822724.
- ↑ "Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization". The Journal of Neuroscience 29 (4): 1175–90. January 2009. doi:10.1523/JNEUROSCI.3328-08.2009. PMID 19176826.
- ↑ "A parcellation scheme for human left lateral parietal cortex". Neuron 67 (1): 156–70. July 2010. doi:10.1016/j.neuron.2010.05.025. PMID 20624599.
- ↑ "The connectional organization of the cortico-thalamic system of the cat". Cerebral Cortex 9 (3): 277–99. 1999. doi:10.1093/cercor/9.3.277. PMID 10355908.
- ↑ "Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database". Neuroinformatics 2 (2): 127–44. 2004. doi:10.1385/NI:2:2:127. PMID 15319511.
- ↑ "The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network". Nature Reviews. Neuroscience 10 (4): 272–82. April 2009. doi:10.1038/nrn2614. PMID 19300446. https://zenodo.org/record/3426276.
- ↑ "Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain". The Journal of Comparative Neurology 513 (5): 532–41. April 2009. doi:10.1002/cne.21974. PMID 19226510.
- ↑ 17.0 17.1 "The WU-Minn Human Connectome Project: an overview". NeuroImage 80: 62–79. October 2013. doi:10.1016/j.neuroimage.2013.05.041. PMID 23684880.
- ↑ 18.0 18.1 "Sequencing the connectome". PLOS Biology 10 (10): e1001411. 2012. doi:10.1371/journal.pbio.1001411. PMID 23109909.

- ↑ 19.0 19.1 "The structure of the nervous system of the nematode Caenorhabditis elegans". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 314 (1165): 1–340. November 1986. doi:10.1098/rstb.1986.0056. PMID 22462104. Bibcode: 1986RSPTB.314....1W.
- ↑ "Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system". Nature 450 (7166): 56–62. November 2007. doi:10.1038/nature06293. PMID 17972876. Bibcode: 2007Natur.450...56L.
- ↑ "Ome sweet ome: what can the genome tell us about the connectome?". Current Opinion in Neurobiology 18 (3): 346–53. June 2008. doi:10.1016/j.conb.2008.08.010. PMID 18801435.
- ↑ "A technicolour approach to the connectome". Nature Reviews. Neuroscience 9 (6): 417–22. June 2008. doi:10.1038/nrn2391. PMID 18446160.
- ↑ 23.0 23.1 "Network anatomy and in vivo physiology of visual cortical neurons". Nature 471 (7337): 177–82. March 2011. doi:10.1038/nature09802. PMID 21390124. Bibcode: 2011Natur.471..177B.
- ↑ 24.0 24.1 "Wiring specificity in the direction-selectivity circuit of the retina". Nature 471 (7337): 183–8. March 2011. doi:10.1038/nature09818. PMID 21390125. Bibcode: 2011Natur.471..183B.
- ↑ "Functional Clusters, Hubs, and Communities in the Cortical Microconnectome". Cerebral Cortex 25 (10): 3743–57. October 2015. doi:10.1093/cercor/bhu252. PMID 25336598.
- ↑ "Staining and embedding the whole mouse brain for electron microscopy". Nature Methods 9 (12): 1198–201. December 2012. doi:10.1038/nmeth.2213. PMID 23085613.
- ↑ Cepelewicz, Jordana (March 8, 2016). "The U.S. Government Launches a $100-Million "Apollo Project of the Brain"". Scientific American. Springer Nature America. http://www.scientificamerican.com/article/the-u-s-government-launches-a-100-million-apollo-project-of-the-brain/.
- ↑ Emily, Singer (April 6, 2016). "Mapping the Brain to Build Better Machines". Simons Foundation. https://www.quantamagazine.org/mapping-the-brain-to-build-better-machines-20160406/.
- ↑ "Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization". Journal of Neurophysiology 103 (1): 297–321. January 2010. doi:10.1152/jn.00783.2009. PMID 19889849.
- ↑ "Consciousness, plasticity, and connectomics: the role of intersubjectivity in human cognition". Frontiers in Psychology 2: 20. 2011. doi:10.3389/fpsyg.2011.00020. PMID 21687435.
- ↑ "Exploring the network dynamics underlying brain activity during rest". Progress in Neurobiology 114: 102–31. March 2014. doi:10.1016/j.pneurobio.2013.12.005. PMID 24389385.
- ↑ "Network structure of cerebral cortex shapes functional connectivity on multiple time scales". Proceedings of the National Academy of Sciences of the United States of America 104 (24): 10240–5. June 2007. doi:10.1073/pnas.0701519104. PMID 17548818. Bibcode: 2007PNAS..10410240H.
- ↑ "Modeling the outcome of structural disconnection on resting-state functional connectivity". NeuroImage 62 (3): 1342–53. September 2012. doi:10.1016/j.neuroimage.2012.06.007. PMID 22705375.
- ↑ "Dynamical consequences of lesions in cortical networks". Human Brain Mapping 29 (7): 802–9. July 2008. doi:10.1002/hbm.20579. PMID 18438885.
- ↑ "Studying the human brain anatomical network via diffusion-weighted MRI and Graph Theory". NeuroImage 40 (3): 1064–76. April 2008. doi:10.1016/j.neuroimage.2007.10.060. PMID 18272400.
- ↑ "MR diffusion tensor spectroscopy and imaging". Biophysical Journal 66 (1): 259–67. January 1994. doi:10.1016/S0006-3495(94)80775-1. PMID 8130344. Bibcode: 1994BpJ....66..259B.
- ↑ "Estimation of the effective self-diffusion tensor from the NMR spin echo". Journal of Magnetic Resonance, Series B 103 (3): 247–54. March 1994. doi:10.1006/jmrb.1994.1037. PMID 8019776. Bibcode: 1994JMRB..103..247B. https://zenodo.org/record/1229906.
- ↑ "Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography". Cerebral Cortex 19 (3): 524–36. March 2009. doi:10.1093/cercor/bhn102. PMID 18567609.
- ↑ 39.0 39.1 "Mapping human whole-brain structural networks with diffusion MRI". PLOS ONE 2 (7): e597. July 2007. doi:10.1371/journal.pone.0000597. PMID 17611629. Bibcode: 2007PLoSO...2..597H.

- ↑ "Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging". Magnetic Resonance in Medicine 54 (6): 1377–86. December 2005. doi:10.1002/mrm.20642. PMID 16247738.
- ↑ "Imagerie de diffusion in vivo par résonance magnétique nucléaire" (in fr). Comptes Rendus de l'Académie des Sciences 93 (5): 27–34. 1985.
- ↑ "MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders". Radiology 161 (2): 401–7. November 1986. doi:10.1148/radiology.161.2.3763909. PMID 3763909.
- ↑ "Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers". NeuroImage 41 (4): 1267–77. July 2008. doi:10.1016/j.neuroimage.2008.03.036. PMID 18495497.
- ↑ "Small-world connectivity, motif composition, and complexity of fractal neuronal connections". Bio Systems 85 (1): 55–64. July 2006. doi:10.1016/j.biosystems.2006.02.008. PMID 16757100.
- ↑ "Identification and classification of hubs in brain networks". PLOS ONE 2 (10): e1049. October 2007. doi:10.1371/journal.pone.0001049. PMID 17940613. Bibcode: 2007PLoSO...2.1049S.

- ↑ "Mapping the structural core of human cerebral cortex". PLOS Biology 6 (7): e159. July 2008. doi:10.1371/journal.pbio.0060159. PMID 18597554.

- ↑ "How to Direct the Edges of the Connectomes: Dynamics of the Consensus Connectomes and the Development of the Connections in the Human Brain". PLOS ONE 11 (6): e0158680. 2016. doi:10.1371/journal.pone.0158680. PMID 27362431. Bibcode: 2016PLoSO..1158680K.
- ↑ 48.0 48.1 Crimi, Alessandro; Giancardo, Luca; Sambataro, Fabio; Diego, Sona (2019). "MultiLink analysis: brain network comparison via sparse connectivity analysis". Scientific Reports 9 (1): 1–13. doi:10.1038/s41598-018-37300-4. PMID 30635604. Bibcode: 2019NatSR...9...65C.
- ↑ Zalesky, Andrew; Fornito, Alex; Bullmore, Edward (2010). "Network-based statistic: identifying differences in brain networks.". NeuroImage 53 (4): 1197–1207. doi:10.1016/j.neuroimage.2010.06.041. PMID 20600983.
- ↑ "Sex differences in the structural connectome of the human brain". Proceedings of the National Academy of Sciences of the United States of America 111 (2): 823–8. January 2014. doi:10.1073/pnas.1316909110. PMID 24297904. Bibcode: 2014PNAS..111..823I.
- ↑ "Graph Theoretical Analysis Reveals: Women's Brains Are Better Connected than Men's". PLOS ONE 10 (7): e0130045. 2015. doi:10.1371/journal.pone.0130045. PMID 26132764. Bibcode: 2015PLoSO..1030045S.
- ↑ "Brain size bias compensated graph-theoretical parameters are also better in women's structural connectomes". Brain Imaging and Behavior 12 (3): 663–673. April 2017. doi:10.1007/s11682-017-9720-0. PMID 28447246.
- ↑ "Age- and gender-related differences in the cortical anatomical network". The Journal of Neuroscience 29 (50): 15684–93. December 2009. doi:10.1523/JNEUROSCI.2308-09.2009. PMID 20016083.
- ↑ "Circular representation of human cortical networks for subject and population-level connectomic visualization". NeuroImage 60 (2): 1340–51. April 2012. doi:10.1016/j.neuroimage.2012.01.107. PMID 22305988.
- ↑ "Patient-tailored connectomics visualization for the assessment of white matter atrophy in traumatic brain injury". Frontiers in Neurology 3: 10. 2012. doi:10.3389/fneur.2012.00010. PMID 22363313.
- ↑ "Mapping connectivity damage in the case of Phineas Gage". PLOS ONE 7 (5): e37454. 2012. doi:10.1371/journal.pone.0037454. PMID 22616011. Bibcode: 2012PLoSO...737454V.

- ↑ "Topological impact of negative links on the stability of resting-state brain network". Scientific Reports 11 (1): 2176. January 2021. doi:10.1038/s41598-021-81767-7. PMID 33500525. Bibcode: 2021NatSR..11.2176S.
- ↑ Greenough, William T.; Bailey, Craig H. (January 1988). "The anatomy of a memory: convergence of results across a diversity of tests". Trends in Neurosciences 11 (4): 142–147. doi:10.1016/0166-2236(88)90139-7.
- ↑ 59.0 59.1 "Rewiring the connectome: Evidence and effects". Neuroscience and Biobehavioral Reviews 88: 51–62. May 2018. doi:10.1016/j.neubiorev.2018.03.001. PMID 29540321.
- ↑ 60.0 60.1 Witvliet, Daniel; Mulcahy, Ben; Mitchell, James K.; Meirovitch, Yaron; Berger, Daniel R.; Wu, Yuelong; Liu, Yufang; Koh, Wan Xian et al. (August 2021). "Connectomes across development reveal principles of brain maturation" (in en). Nature 596 (7871): 257–261. doi:10.1038/s41586-021-03778-8. ISSN 1476-4687. PMID 34349261. Bibcode: 2021Natur.596..257W.
- ↑ 61.0 61.1 Meirovitch, Yaron; Kang, Kai; Draft, Ryan W.; Pavarino, Elisa C.; Henao E., Maria F.; Yang, Fuming; Turney, Stephen G.; Berger, Daniel R. et al. (September 2021). "Neuromuscular connectomes across development reveal synaptic ordering rules" (in en). bioRxiv. doi:10.1101/2021.09.20.460480.
- ↑ "Extension of corticocortical afferents into the anterior bank of the intraparietal sulcus by tool-use training in adult monkeys". Neuropsychologia 44 (13): 2636–46. 2006. doi:10.1016/j.neuropsychologia.2005.11.020. PMID 16427666.
- ↑ "Axonal sprouting and formation of terminals in the adult cerebellum during associative motor learning". The Journal of Neuroscience 33 (45): 17897–907. November 2013. doi:10.1523/JNEUROSCI.0511-13.2013. PMID 24198378.
- ↑ "The emergence of functional microcircuits in visual cortex". Nature 496 (7443): 96–100. April 2013. doi:10.1038/nature12015. PMID 23552948. Bibcode: 2013Natur.496...96K.
- ↑ "Experience-dependent and cell-type-specific spine growth in the neocortex" (in En). Nature 441 (7096): 979–83. June 2006. doi:10.1038/nature04783. PMID 16791195. Bibcode: 2006Natur.441..979H.
- ↑ "Spine growth precedes synapse formation in the adult neocortex in vivo" (in En). Nature Neuroscience 9 (9): 1117–24. September 2006. doi:10.1038/nn1747. PMID 16892056.
- ↑ 67.0 67.1 "Rapid formation and selective stabilization of synapses for enduring motor memories" (in En). Nature 462 (7275): 915–9. December 2009. doi:10.1038/nature08389. PMID 19946267. Bibcode: 2009Natur.462..915X.
- ↑ Jensen, K. Thor (8 May 2020). "Will We Ever Be Able to Upload Our Brains?". https://www.pcmag.com/news/will-we-ever-be-able-to-upload-our-brains.
- ↑ Brouillette, Monique (21 April 2022). "Mapping the brain to understand the mind" (in en). Knowable Magazine | Annual Reviews. doi:10.1146/knowable-042122-1. https://knowablemagazine.org/article/mind/2022/mapping-brain-understand-mind.
- ↑ "Ay's Neuroanatomy of C. elegans for Computation" (in en). https://www.crcpress.com/Ays-Neuroanatomy-of-C-Elegans-for-Computation/Achacoso-Yamamoto/p/book/9780849342349.
- ↑ Yamamoto, William S.; Achacoso, Theodore B. (1992-06-01). "Scaling up the nervous system of Caenorhabditis elegans: Is one ape equal to 33 million worms?". Computers and Biomedical Research 25 (3): 279–291. doi:10.1016/0010-4809(92)90043-A. ISSN 0010-4809. PMID 1611892.
- ↑ Achacoso, Theodore B.; Fernandez, Victor; Nguyen, Duc C.; Yamamoto, William S. (1989-11-08). "Computer Representation of the Synaptic Connectivity of Caenorhabditis Elegans". Proceedings of the Annual Symposium on Computer Application in Medical Care: 330–334. ISSN 0195-4210.
- ↑ "Structural properties of the Caenorhabditis elegans neuronal network". PLOS Computational Biology 7 (2): e1001066. February 2011. doi:10.1371/journal.pcbi.1001066. PMID 21304930. Bibcode: 2011PLSCB...7E1066V.

- ↑ Cook, Steven J.; Jarrell, Travis A.; Brittin, Christopher A.; Wang, Yi; Bloniarz, Adam E.; Yakovlev, Maksim A.; Nguyen, Ken C. Q.; Tang, Leo T.-H. et al. (3 July 2019). "Whole-animal connectomes of both Caenorhabditis elegans sexes". Nature 571 (7763): 63–71. doi:10.1038/s41586-019-1352-7. PMID 31270481. Bibcode: 2019Natur.571...63C.
- ↑ "A community-developed open-source computational ecosystem for big neuro data". Nature Methods 15 (11): 846–847. October 2018. doi:10.1038/s41592-018-0181-1. PMID 30377345. Bibcode: 2018arXiv180402835B.
- ↑ Witvliet, Daniel; Mulcahy, Ben; Mitchell, James K.; Meirovitch, Yaron; Berger, Daniel R.; Wu, Yuelong; Liu, Yufang; Koh, Wan Xian et al. (2020-04-30) (in en), Connectomes across development reveal principles of brain maturation in C. elegans, doi:10.1101/2020.04.30.066209, https://www.biorxiv.org/content/10.1101/2020.04.30.066209v1, retrieved 2024-01-23, "About 43% of all connections and 16% of all synapses were not conserved between animals. This degree of variability contrasts with the widely held view that the C. elegans connectome is hardwired."
- ↑ Cook, Steven J.; Kalinski, Cristine A.; Hobert, Oliver (2023-06-05). "Neuronal contact predicts connectivity in the C. elegans brain". Current Biology 33 (11): 2315–2320.e2. doi:10.1016/j.cub.2023.04.071. ISSN 0960-9822. PMID 37236179. https://www.sciencedirect.com/science/article/pii/S0960982223005535. "C. elegans neurons show inter-individual variability in adjacency and connectivity".
- ↑ Schlegel, Philipp (2021-05-25). "Information flow, cell types and stereotypy in a full olfactory connectome". eLife 10 (10). doi:10.7554/eLife.66018. PMID 34032214.
- ↑ Winding, Michael; Pedigo, Benjamin; Barnes, Christopher; Patsolic, Heather; Park, Youngser; Kazimiers, Tom; Fushiki, Akira; Andrade, Ingrid et al. (10 Mar 2023). "The connectome of an insect brain". Science 379 (6636): eadd9330. doi:10.1126/science.add9330. PMID 36893230.
- ↑ Rosen, Jill (2023-03-09). "Scientists complete first map of an insect brain" (in en). https://hub.jhu.edu/2023/03/09/scientists-complete-first-map-of-an-insect-brain/.
- ↑ "First wiring map of insect brain complete" (in en). 2023-03-10. https://www.cam.ac.uk/research/news/first-wiring-map-of-insect-brain-complete.
- ↑ Zheng, Zhihao (2018-07-19). "A complete electron microscopy volume of the brain of adult Drosophila melanogaster.". Cell 174.3 (2018): 730–743. doi:10.1016/j.cell.2018.06.019. PMID 30033368.
- ↑ Scheffer, Louis (2020). "A connectome and analysis of the adult Drosophila central brain". eLife 9 (2020). doi:10.7554/eLife.57443. PMID 32880371.
- ↑ Dorkenwald, Sven (2023-06-29). "Neuronal wiring diagram of an adult brain". bioRxiv. doi:10.1101/2023.06.27.546656. PMID 37425937.
- ↑ Phelps, Jasper (2021-02-04). "Reconstruction of motor control circuits in adult Drosophila using automated transmission electron microscopy". Cell 184 (2021): 759–774. doi:10.1016/j.cell.2020.12.013. PMID 33400916.
- ↑ Takemura, Shin-ya (2023-06-06). "A Connectome of the Male Drosophila Ventral Nerve Cord". bioRxiv. doi:10.1101/2023.06.05.543757.
External links
- Database of hundreds of braingraphs with different resolutions and weight functions at braingraph.org
- The NIH Blueprint for Neuroscience Research
- TED talk by Sebastian Seung: I am my connectome
- MITK Diffusion: Free software for the processing of diffusion-weighted MR data including connectomics
 |
 KSF
KSF