PyMOL
Topic: Software
 From HandWiki - Reading time: 4 min
From HandWiki - Reading time: 4 min
 | |
 A PyMOL instance, with the Viewer and GUI visible. | |
| Original author(s) | Warren Lyford DeLano |
|---|---|
| Developer(s) | Schrödinger, Inc. |
| Initial release | 2000 |
| Stable release | 2.5.5[1]
/ 17 April 2023 |
| Written in | C, C++, Python |
| Operating system | Cross-platform: macOS, Unix, Linux, Windows |
| Platform | IA-32, x86-64 |
| Available in | English |
| Type | Molecular modelling |
| License | Originally the Python License,[2] now proprietary[3] |
| Website | pymol |
PyMOL is an open source but proprietary[3] molecular visualization system created by Warren Lyford DeLano. It was commercialized initially by DeLano Scientific LLC, which was a private software company dedicated to creating useful tools that become universally accessible to scientific and educational communities. It is currently commercialized by Schrödinger, Inc. As the original software license was a permissive licence, they were able to remove it; new versions are no longer released under the Python license, but under a custom license (granting broad use, redistribution, and modification rights, but assigning copyright to any version to Schrodinger, LLC.),[3] and some of the source code is no longer released.[4] PyMOL can produce high-quality 3D images of small molecules and biological macromolecules, such as proteins. According to the original author, by 2009, almost a quarter of all published images of 3D protein structures in the scientific literature were made using PyMOL.[citation needed]
PyMOL is one of the few mostly open-source model visualization tools available for use in structural biology. The Py part of the software's name refers to the program having been written in the programming language Python.
PyMOL uses OpenGL Extension Wrangler Library (GLEW) and FreeGLUT, and can solve Poisson–Boltzmann equations using the Adaptive Poisson Boltzmann Solver.[5] PyMOL used Tk for the GUI widgets and had native Aqua binaries for macOS through Schrödinger, which were replaced with a PyQt user interface on all platforms with the release of version 2.0.[6]
History and commercialization
Early versions of PyMol were released under the Python License. On 1 August 2006, DeLano Scientific adopted a controlled-access download system for precompiled PyMOL builds (including betas) distributed by the company. Access to these executables is now limited to registered users who are paying customers; educational builds are available free to students and teachers. However, most of the current source code continues to be available for free, as are older precompiled builds. While the build systems for other platforms are open, the Windows API (WinAPI, Win32) build system is not, although unofficial Windows binaries are available online.[7] Anyone can either compile an executable from the Python-licensed source code or pay for a subscription to support services to obtain access to precompiled executables.
On 8 January 2010, Schrödinger, Inc. reached an agreement to acquire PyMOL. The firm assumed development, maintenance, support, and sales of PyMOL, including all then-valid subscriptions. They also continue to actively support the PyMOL open-source community. In 2017, Schrödinger revamped the distribution system to unify the user interface under Qt and the package management under Anaconda, and released it as PyMol v2.[6] This version restricts some new functionalities and adds a watermark to the visualization if used unlicensed beyond the 30-day trial period; the overall license policy is similar to the DeLano system. The source code remains mostly available, this time under a BSD-like license.[8] As with the previous distribution, unofficial Windows binaries in the wheel format are available,[7] and indeed Linux distributions continue to provide their own builds of the open-source code.
Element colors
PyMOL applies ball-coloring by element.
| Periodic table using PyMOL element colors as background[9] | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ① | |||||||||||||||||||
| ② | |||||||||||||||||||
| ③ | |||||||||||||||||||
| ④ | |||||||||||||||||||
| ⑤ | |||||||||||||||||||
| ⑥ | |||||||||||||||||||
| ⑦ | |||||||||||||||||||
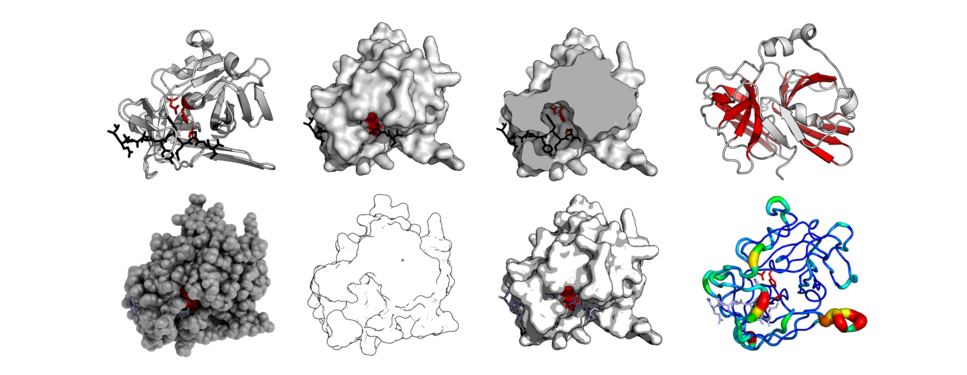
Gallery
The same protein structure (TEV protease - PDB: 1LVB) rendered in different modes. Standard cartoon, surface, cut-through of surface, highlighted barrels, 'QuteMol'-like, 'Goodsell'-like, glossy-surface, and b-factor putty.
See also
- Comparison of software for molecular mechanics modeling
- List of molecular graphics systems
- Molecular modelling
- Abalone
- Gabedit
- Molden
- Molekel
- RasMol
- SAMSON
- UCSF Chimera
- List of free and open-source software packages
References
- ↑ "Download PyMol" (in en). https://pymol.org/2/#download.
- ↑ "PyMOL Molecular Graphics System". SourceForge. http://sourceforge.net/projects/pymol/.
- ↑ 3.0 3.1 3.2 "Open-Source PyMOL". Schrodinger, Inc.. 5 November 2021. https://github.com/schrodinger/pymol-open-source/blob/master/LICENSE.
- ↑ "PyMOL | pymol.org". https://pymol.org/2/. "Open-Source Philosophy
PyMOL is a commercial product, but we make most of its source code freely available under a permissive license. The open source project is maintained by Schrödinger and ultimately funded by everyone who purchases a PyMOL license.
Open source enables open science.
This was the vision of the original PyMOL author Warren L. DeLano." - ↑ "APBS". http://www.poissonboltzmann.org/apbs/.
- ↑ 6.0 6.1 "PyMOL v2.0 Release Notes". https://pymol.org/dokuwiki/?id=media:new2.
- ↑ 7.0 7.1 "Python Extension Packages for Windows - Christoph Gohlke". https://www.lfd.uci.edu/~gohlke/pythonlibs/#pymol.
- ↑ "schrodinger/pymol-open-source". https://github.com/schrodinger/pymol-open-source.
- ↑ "Color Values". pymolwiki. 2010. https://pymolwiki.org/index.php/Color_Values#Chemical_element_colours.
External links
 |
 KSF
KSF