Heme
 From Wikidoc - Reading time: 6 min
From Wikidoc - Reading time: 6 min

|
WikiDoc Resources for Heme |
|
Articles |
|---|
|
Media |
|
Evidence Based Medicine |
|
Clinical Trials |
|
Ongoing Trials on Heme at Clinical Trials.gov Clinical Trials on Heme at Google
|
|
Guidelines / Policies / Govt |
|
US National Guidelines Clearinghouse on Heme
|
|
Books |
|
News |
|
Commentary |
|
Definitions |
|
Patient Resources / Community |
|
Directions to Hospitals Treating Heme Risk calculators and risk factors for Heme
|
|
Healthcare Provider Resources |
|
Continuing Medical Education (CME) |
|
International |
|
|
|
Business |
|
Experimental / Informatics |
Overview[edit | edit source]
A heme or haem is a prosthetic group that consists of an iron atom contained in the center of a large heterocyclic organic ring called a porphyrin. Not all porphyrins contain iron, but a substantial fraction of porphyrin-containing metalloproteins have heme as their prosthetic subunit; these are known as hemoproteins.
Types[edit | edit source]
Major hemes[edit | edit source]
There are several biologically important kinds of heme:
| Heme a | Heme b | Heme c | Heme o | |
|---|---|---|---|---|
| PubChem number | 7888115 | 444098 | 444125 | 6323367 |
| Chemical formula | C49H56O6N4Fe | C34H32O4N4Fe | C34H36O4N4S2Fe | C49H58O5N4Fe |
| Functional group at C3 | Hydroxyfarnesyl | -CH=CH2 | -CH-(CH3)-SH | Hydroxyfarnesyl |
| Functional group at C8 | -CH=CH2 | -CH=CH2 | -CH-(CH3)-SH | -CH=CH2 |
| Functional group at C18 | -CH=O | -CH3 | -CH3 | -CH3 |
The most common type is heme B; other important types include heme A and heme C. Isolated hemes are commonly designated by capital letters while hemes bound to proteins are designated by lower case letters. Cytochrome a refers to the heme A in specific combination with membrane protein forming a portion of cytochrome c oxidase.
Other hemes[edit | edit source]
- Heme L is the derivative of heme B which is covalently attached to the protein of lactoperoxidase, eosinophil peroxidase and thyroid peroxidase. The addition of peroxide with the glutamyl-375 and aspartyl-225 of lactoperoxidase forms ester bonds between these amino acid residues and the heme 1- and 5-methyl groups, respectively. Similar ester bonds with these two methyl groups are thought to form in eosinophil and thyroid peroxidases. Heme L is one important characteristic of animal peroxidases; plant peroxidases incorporate heme B. Lactoperoxidase and eosinophil peroxidase are protective enzymes responsible for the destruction of invading bacteria and virus. Thyroid peroxidase is the enzyme catalyzing the biosynthesis of the important thyroid hormones. Because lactoperoxidase destroys invading organisms in excrement, it is thought to be an important protective enzyme.
- Heme M is the derivative of heme B covalently bound at the active site of myeloperoxidase. Heme M also contains the two ester bonds at the heme 1- and 5-methyls, much as the other mammalian peroxidases. In addition, a unique sulfonium ion linkage between the sulfur of a methionyl aminoacid residue and the heme 2-vinyl group is formed, giving this enzyme the unique capability of easily oxidizing chloride and bromide ions. Myeloperoxidase is present in mammalian neutrophils and is responsible for the destruction of invading bacteria and virus. It also synthesizes hypobromite by "mistake" which is a known mutagenic compound.
- Heme D is another derivative of heme B, but in which the propionic acid side chain at the carbon of position 6, ring III is bound to this carbon both via the usual C-C bond but also by the carboxyl oxygen, giving heme D a fifth ring and a lactone. Ring III is also hydroxylated at position 5, in a conformation trans to the new lactone group. [1] Heme D is the site for oxygen reduction to water of many types of bacteria at low oxygen tension.
- Heme S is related to heme B by the having a formyl group at position 2 in place of the 2-vinyl group. Heme S is found in the hemoglobin of marine worms. The correct structures of heme B and heme S were first elucidated by the great German chemist Hans Fischer.
The names of cytochromes typically (but not always) reflect the kinds of hemes they contain: cytochrome a contains heme A, cytochrome c contains heme C, etc.
Function[edit | edit source]
Hemoproteins have diverse biological functions including the transportation of diatomic gases, chemical catalysis, diatomic gas detection, and electron transfer. The heme iron serves as a source or sink of electrons during electron transfer or redox chemistry. In peroxidase reactions, the porphyrin molecule also serves as an electron source. In the transportation or detection of diatomic gases, the gas binds to the heme iron. During the detection of diatomic gases, the binding of the gas ligand to the heme iron induces conformational changes in the surrounding protein.
It has been speculated that the original evolutionary function of hemoproteins was electron transfer in primitive sulfur-based photosynthesis pathways in ancestral cyanobacteria before the appearance of molecular oxygen. [2]
Hemoproteins achieve their remarkable functional diversity by modifying the environment of the heme macrocycle within the protein matrix. For example, the ability of hemoglobin to effectively deliver oxygen to tissues is due to specific amino acid residues located near the heme molecule. Hemoglobin binds oxygen in the pulmonary vasculature, where the pH is high and the pCO2 is low, and releases it in the tissues, where the situations are reversed. This phenomenon is known as the Bohr effect. The molecular mechanism behind this effect is the steric organisation of the globin chain; a histidine residue, located adjacent to the heme group, becomes positively charged under acid circumstances, sterically releasing oxygen from the heme group.
Synthesis[edit | edit source]
Details of heme synthesis can be found in the article on porphyrin.
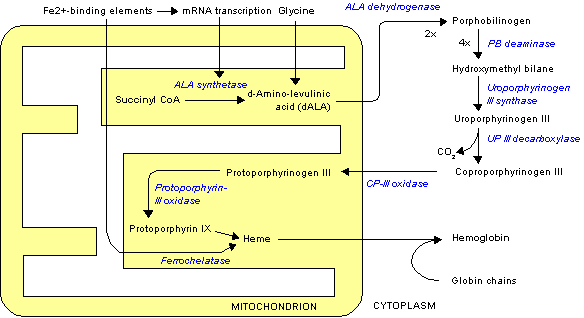
The enzymatic process that produces heme is properly called porphyrin synthesis, as all the intermediates are tetrapyrroles that are chemically classified are porphyrins. The process is highly conserved across biology. In humans, this pathway serves almost exclusively to form heme. In other species, it also produces similar substances such as cobalamin (vitamin B12).
The pathway is initiated by the synthesis of D-Aminolevulinic acid (dALA or δALA) from the amino acid glycine and succinyl-CoA from the citric acid cycle (Krebs cycle). The rate-limiting enzyme responsible for this reaction, ALA synthase, is strictly regulated by intracellular iron levels and heme concentration. A low-iron level, e.g., in iron deficiency, leads to decreased porphyrin synthesis, which prevents accumulation of the toxic intermediates. This mechanism is of therapeutic importance: infusion of heme arginate of hematin can abort attacks of porphyria in patients with an inborn error of metabolism of this process, by reducing transcription of ALA synthase.
The organs mainly involved in heme synthesis are the liver and the bone marrow, although every cell requires heme to function properly. Heme is seen as an intermediate molecule in catabolism of haemoglobin in the process of bilirubin metabolism.
Degradation[edit | edit source]
In the first step, heme is converted to biliverdin by the enzyme heme oxygenase (HOXG). NADPH is used as the reducing agent, molecular oxygen enters the reaction, carbon monoxide is produced and the iron is released from the molecule as the ferric ion (Fe3+).
HOXG
heme --------------> biliverdin + Fe3+
/ \
H+ + NADPH NADP+
O2 CO
In the second reaction, biliverdin is converted to bilirubin by biliverdin reductase (BVR):
BVR
biliverdin -----------> bilirubin
/ \
H+ + NADPH NADP+
Bilirubin is transported into the liver bound to a protein (serum albumin), where it is conjugated with glucuronic acid to become more water soluble. The reaction is catalyzed by the enzyme UDP-glucuronide transferase (UDPGUTF).
UDPGUTF
bilirubin + 2 UDP-glucuronate ------------> bilirubin diglucuronide
\
2 UMP + 2 Pi
This form of bilirubin is excreted from the liver in bile. The intestinal bacteria deconjugate bilirubin diglucuronide and convert bilirubin to urobilinogens. Some urobilinogen is absorbed by intestinal cells and transported into the kidneys and excreted with urine. The remainder travels down the digestive tract and is excreted as stercobilinogen, which is responsible for the color of feces.
Genes[edit | edit source]
The following genes are part of the chemical pathway for making heme:
- ALAD: aminolevulinic acid, delta-, dehydratase
- ALAS1: aminolevulinate, delta-, synthase 1
- ALAS2: aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia)
- CPOX: coproporphyrinogen oxidase
- FECH: ferrochelatase (protoporphyria)
- HMBS: hydroxymethylbilane synthase
- PPOX: protoporphyrinogen oxidase
- UROD: uroporphyrinogen decarboxylase
- UROS: uroporphyrinogen III synthase (congenital erythropoietic porphyria)
See also[edit | edit source]
References[edit | edit source]
- ↑ Timkovich, R., Cork, M.S., Gennis, R.B. and Johnson, P.Y. (1985). "Proposed Structure of Heme d, a Prostetic Group of Bacterial Terminal Oxidases". Journal of the American Chemical Society. 107 (21): 6069–6075.
- ↑ Hardison, R. (1999). "The Evolution of Hemoglobin Studies: of a very ancient protein suggest that changes in gene regulation are an important part of the evolutionary story". American Scientist. 87 (2): 126.
de:Häm eo:Hemo io:Hemeo it:Eme he:הם hu:Hem nl:Heemverbinding nn:Heme sl:Hem fi:Hemi (kemia) sv:Hemgrupp
 KSF
KSF