Initiator element gene transcription laboratory
 From Wikidoc - Reading time: 44 min
From Wikidoc - Reading time: 44 min
Editor-In-Chief: Henry A. Hoff
{{free media}}A laboratory is a specialized activity, a construct, you create where you as a student, teacher, or researcher can have hands-on, or as close to hands-on as possible, experience actively analyzing an entity, source, or object of interest. Usually, there's more to do than just analyzing. The construct is often a room, building or institution equipped for scientific research, experimentation as well as analysis.
This laboratory is a continuation of the previous laboratory.
In the room next door is an astronaut on the Mars expedition, three months along on the six-month journey. A physician and lab assistants have been performing tests on her. Because she has been in zero gravity for more than three months her body chemistry and anatomy now differ from what it was in the controlled gravity environment of Earth. She has lost about 10 % each of her bone, muscle, and brain mass. Comparisons with gene expression sequences now and when on Earth have found that the gene expression for alpha-1-B glycoprotein is not normal. If a way to correct this expression cannot be found she must be returned to Earth maybe to recover, maybe not!
But, it is unlikely she will survive three more months at zero g either to be returned to Earth or put on Mars. Worse, the microgravity may not be the only culprit. There is also the radiation of the interplanetary medium.
You have been tasked to examine her DNA to confirm, especially with the extended data between ZNF497 and A1BG, the presence or absence of initiator elements regarding the possible expression of alpha-1-B glycoprotein.
Consensus sequences[edit | edit source]
As in other metazoans, for genes lacking a TATA box, the Inr is functionally analogous, with a base pair (bp) consensus 5'-YYA+1NWYY-3', to direct transcription initiation.[1] Using the degenerate nucleotide code, the consensus sequence is 5'-C/T-C/T-A-A/C/G/T-A/T-C/T-C/T-3', or in the direction of transcription on the template strand: 3'-C/T-C/T-A-A/C/G/T-A/T-C/T-C/T-5'.
"TATA-less core promoters that lack AT-rich sequences in the -30 region and do not stably bind TBP are likely to assemble PICs via alternative pathways and to be regulated by distinct mechanisms (Smale and Kadonaga, 2003). However, the number of such bona fide TATA-less genes remains unclear in eukaryotic genomes."[2]
In Entamoeba histolytica, the consensus sequence is AAAAATTCA.[3]
The Inr has the consensus sequence YYANWYY.[4] Similarly to the TATA box, the Inr element facilitates the binding of transcription Factor II D (TATA binding protein TAF).[4]
Nucleotides[edit | edit source]
DNA mapping has been performed. Her DNA for A1BG promoters can be found at Gene_transcriptions/A1BG#Nucleotides.
Programming[edit | edit source]
Sample programs for preparing test programs are available at Gene transcriptions/A1BG/Programming.
Hypotheses[edit | edit source]
- A1BG is not transcribed by an initiator element.
- A1BG is not transcribed by a TATA box.
Core promoters[edit | edit source]
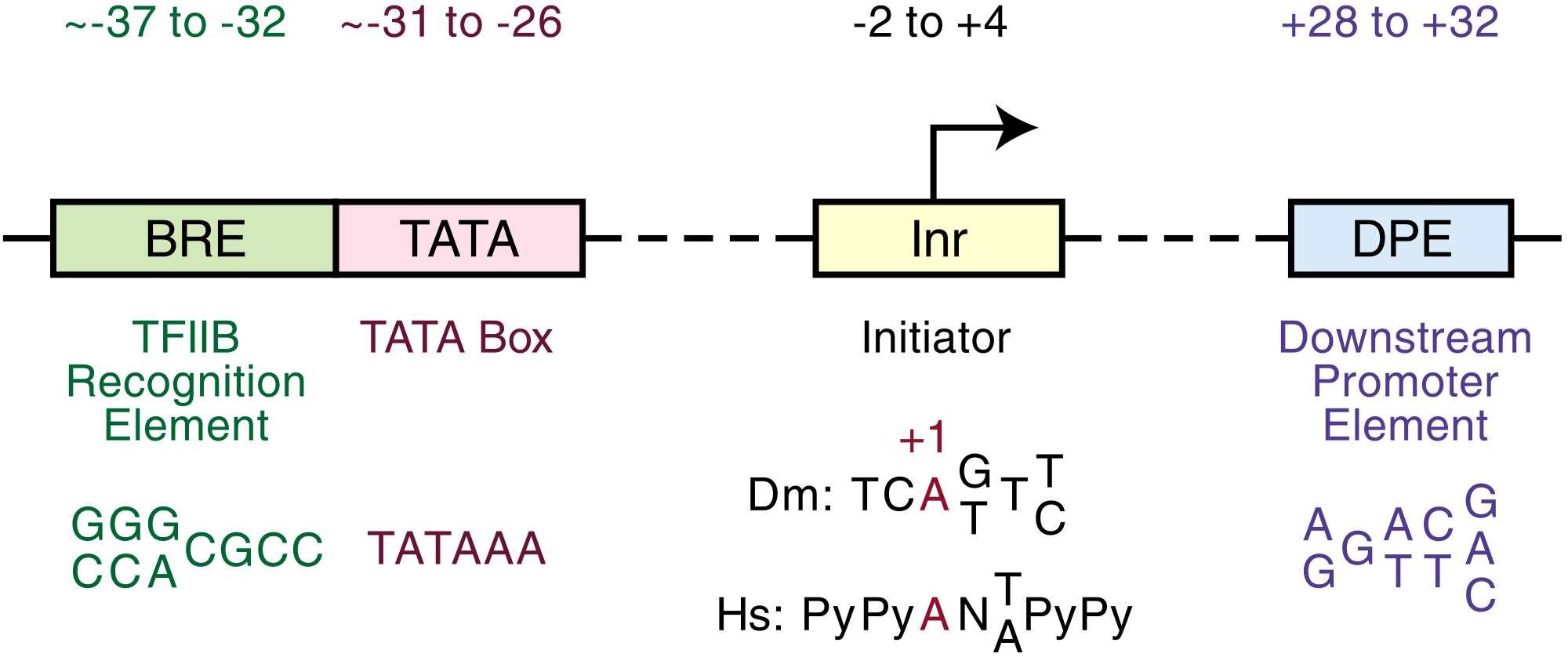
{{free media}}{{fairuse}}The core promoter is approximately -34 nts upstream from the TSS.
From the first nucleotide just after ZSCAN22 to the first nucleotide just before A1BG are 4460 nucleotides. The core promoter on this side of A1BG extends from approximately 4425 to the possible transcription start site at nucleotide number 4460.
To extend the analysis from inside and just on the other side of ZNF497 some 3340 nts have been added to the data. This would place the core promoter some 3340 nts further away from the other side of ZNF497. The TSS would be at about 4300 nts with the core promoter starting at 4266.
Def. "the factors, including RNA polymerase II itself, that are minimally essential for transcription in vitro from an isolated core promoter" is called the basal machinery, or basal transcription machinery.[6]
"The core promoter in human genes is the region from −40 to +40 and flanks the transcription start site (TSS) at +1. Although no single core promoter element is contained in all human promoters, many contain one or more of the following core elements [...]: the TATA box, initiator (Inr), TFIIB recognition elements (BREu and BREd), polypyrimidine initiator (TCT), motif ten element (MTE), and downstream core promoter element (DPE) [...]. Of these, the Inr element encompasses the TSS and is thought to be the most common core promoter element, with previous studies estimating that ∼50% of human core promoters contain an Inr (Gershenzon and Ioshikhes 2005; Yang et al. 2007). The commonly used consensus sequence for the human Inr, which was derived from mutational analyses, is YYANWYY from −2 to +5 (where, Y = C/T, W = A/T, N=A/C/G/T, and +1 is [A)] (Javahery et al. 1994; Lo and Smale 1996)."[7]
"Kadonaga and colleagues (Vo ngoc et al. 2017) devised and implemented a novel multistep approach that combines experimental and computational methods to reinvestigate the human Inr consensus sequence. First, they generated two 5′-GRO-seq (5′ end-selected global run-on followed by sequencing) libraries with human MCF-7 cells to identify the 5′ ends of nascent capped transcripts. Second, they developed a peak-calling algorithm named FocusTSS to find transcripts in the 5′-GRO-seq data sets that were initiated at a focused position on the genome, hence identifying clear TSSs to enable analysis of Inr sequences. FocusTSS identified 7678 TSSs that were in both data sets. Third, to identify sequence motifs enriched among the focused TSSs, they used the HOMER motif discovery tool (Heinz et al. 2010), which yielded an Inr-like consensus sequence of BBCABW from −3 to +3 (where, B = C/G/T, W = A/T, and +1 is [A]). Forty percent of the focused TSSs contained a perfect match to the BBCABW consensus Inr."[7]
The second image down on the right shows relative "locations of select human core promoter elements and the Inr consensus sequence found in promoters with focused TSSs. The promoter elements depicted include BREu (the upstream TFIIB recognition element), TATA (the TATA box), BREd (the downstream TFIIB recognition element), Inr (new consensus sequence shown), MTE, and DPE."[7]
Proximal promoters[edit | edit source]
Def. a "promoter region [juxtaposed to the core promoter that] binds transcription factors that modify the affinity of the core promoter for RNA polymerase.[12][13]"[8] is called a proximal promoter.
The proximal sequence upstream of the gene that tends to contain primary regulatory elements is a proximal promoter.
It is approximately 250 base pairs or nucleotides, nts, upstream of the transcription start site.
The proximal promoter begins about nucleotide number 4210 in the negative direction.
The proximal promoter begins about nucleotide number 4195 in the positive direction.
Distal promoters[edit | edit source]
The "upstream regions of the human [cytochrome P450 family 11 subfamily A] CYP11A and bovine CYP11B genes [have] a distal promoter in each gene. The distal promoters are located at −1.8 to −1.5 kb in the upstream region of the CYP11A gene and −1.5 to −1.1 kb in the upstream region of the CYP11B gene."[9]
"Using cloned chicken βA-globin genes, either individually or within the natural chromosomal locus, enhancer-dependent transcription is achieved in vitro at a distance of 2 kb with developmentally staged erythroid extracts. This occurs by promoter derepression and is critically dependent upon DNA topology. In the presence of the enhancer, genes must exist in a supercoiled conformation to be actively transcribed, whereas relaxed or linear templates are inactive. Distal protein–protein interactions in vitro may be favored on supercoiled DNA because of topological constraints."[10]
Distal promoter regions may be a relatively small number of nucleotides, fairly close to the TSS such as (-253 to -54)[11] or several regions of different lengths, many nucleotides away, such as (-2732 to -2600) and (-2830 to -2800).[12]
The "[d]istal promoter is not a spacer element."[13]
Using an estimate of 2 knts, a distal promoter to A1BG would be expected after nucleotide number 2460.
Any transcription factors before A1BG from the direction of ZN497 may be out to 2300 nts.
Samplings[edit | edit source]
Regarding hypothesis 1[edit | edit source]
Initiator elements[edit | edit source]
For the Basic programs testing consensus sequence YYANWYY (starting with SuccessablesInr.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesInr--.bas, looking for 3'-C/T-C/T-A/G-A/C/G/T-A/T-C/T-C/T-5', 121, 3'-TTGTTCC-5', 71, 3'-CTATACC-5', 77, 3'-CCGTTTC-5', 93, 3'-CCGTACT-5', 124, 3'-CCATATT-5', 181, 3'-CTACATT-5', 247, 3'-TTGGTCC-5', 262, 3'-TTATACT-5', 274, 3'-TCACTCT-5', 301, 3'-CTGCTTT-5', 312, 3'-CCGGTTC-5', 419, 3'-CCAGTCC-5', 441, 3'-TCGGACC-5', 459, 3'-TTGTATC-5', 468, 3'-TCACTTT-5', 473, 3'-TCGGACC-5', 508, 3'-CCGGTTC-5', 556, 3'-CCAGTCC-5', 578, 3'-TTATACC-5', 605, 3'-CCGGTCC-5', 648, 3'-CCGGTTC-5', 692, 3'-CCAGTCC-5', 714, 3'-TCGGACT-5', 732, 3'-TCGCACC-5', 741, 3'-CTACACC-5', 787, 3'-TCGGTTC-5', 874, 3'-TCGGACC-5', 899, 3'-TCGCTCT-5', 913, 3'-TCGGTCC-5', 948, 3'-CCGTACC-5', 953, 3'-TTAGTCC-5', 984, 3'-TTGGACC-5', 1015, 3'-TCACTCT-5', 1079, 3'-TCGGACC-5', 1198, 3'-TTGTACC-5', 1207, 3'-CCACTTT-5', 1212, 3'-CCGCACC-5', 1244, 3'-TTGGATC-5', 1306, 3'-TCAGACC-5', 1356, 3'-TTATTCT-5', 1365, 3'-TCGTTTT-5', 1371, 3'-TTGTTTT-5', 1394, 3'-CCACACT-5', 1479, 3'-TTGCTTC-5', 1555, 3'-CCGTTTT-5', 1561, 3'-TTACTTT-5', 1582, 3'-TTGGATT-5', 1591, 3'-TTAATTT-5', 1697, 3'-TTATACC-5', 1742, 3'-CCGCACC-5', 1897, 3'-CCGTACT-5', 1953, 3'-TTGGACC-5', 1959, 3'-TCGGACC-5', 2009, 3'-TCGTTCT-5', 2023, 3'-TTACACC-5', 2065, 3'-CCGGTCC-5', 2077, 3'-TCACATT-5', 2087, 3'-TCAAACT-5', 2141, 3'-TTGTACC-5', 2152, 3'-CCGCTTT-5', 2157, 3'-CCAGTCC-5', 2250, 3'-TCAAACT-5', 2257, 3'-TCGGACC-5', 2268, 3'-TCGTACC-5', 2277, 3'-CCACTTT-5', 2282, 3'-TTGGACC-5', 2385, 3'-TCGGACC-5', 2435, 3'-TCACTCT-5', 2449, 3'-TCGTTTT-5', 2476, 3'-TTGTTTT-5', 2490, 3'-TCATTCT-5', 2503, 3'-CCGGTCC-5', 2519, 3'-CCAGTCC-5', 2587, 3'-TCACACC-5', 2605, 3'-TTGTACC-5', 2614, 3'-CCACTTT-5', 2619, 3'-TCACACC-5', 2658, 3'-TTGGACC-5', 2720, 3'-TCGGACC-5', 2770, 3'-TCGTACT-5', 2784, 3'-TTGATTC-5', 2914, 3'-CCGATTT-5', 3009, 3'-TTGATTC-5', 3031, 3'-CCGCACC-5', 3047, 3'-TCGGACC-5', 3128, 3'-TTGTTCC-5', 3141, 3'-CCACTTT-5', 3146, 3'-TTGTATT-5', 3169, 3'-CCACACC-5', 3186, 3'-TCGGTTC-5', 3273, 3'-TCGGACC-5', 3298, 3'-TTGTTCT-5', 3307, 3'-TCGTTTT-5', 3313, 3'-TTGTTCT-5', 3340, 3'-TCGTTCT-5', 3374, 3'-CCGAACT-5', 3401, 3'-CCGTATC-5', 3446, 3'-TTGATCT-5', 3463, 3'-TTGGTCT-5', 3486, 3'-CTGTTCT-5', 3759, 3'-CTACACC-5', 3810, 3'-CTGGTCC-5', 3871, 3'-TCATTCT-5', 3893, 3'-CTACTTT-5', 3922, 3'-CCGGTCC-5', 3951, 3'-TCGGACC-5', 4037, 3'-TTGTATC-5', 4046, 3'-TCACTCT-5', 4051, 3'-TTACACT-5', 4092, 3'-CCGGTCC-5', 4102, 3'-CCGTACC-5', 4107, 3'-CCGGTCC-5', 4170, 3'-TCGAACC-5', 4188, 3'-TCACTCT-5', 4202, 3'-TCGGTCT-5', 4233, 3'-CTGCACC-5', 4238, 3'-TCGGACC-5', 4300, 3'-CCAGTTT-5', 4309, 3'-TCGGACC-5', 4349, 3'-TCACACT-5', 4361, 3'-TTACTCC-5', 4557,
- negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesInr-+.bas, looking for 3'-C/T-C/T-A/G-A/C/G/T-A/T-C/T-C/T-5', 45, 3'-TTGTATT-5', 115, 3'-CTGTTTT-5', 147, 3'-CCACACT-5', 345, 3'-CCGGACT-5', 746, 3'-CTGCACT-5', 1372, 3'-CTGCACT-5', 1472, 3'-CCAGACT-5', 1744, 3'-CCACTTC-5', 1914, 3'-CTATTTC-5', 1978, 3'-CCAGTCC-5', 2026, 3'-TCGCTTC-5', 2095, 3'-TCATATT-5', 2178, 3'-CTGCATT-5', 2206, 3'-CCAGATC-5', 2230, 3'-TCAATCT-5', 2235, 3'-CTGTTTC-5', 2263, 3'-TCACTCT-5', 2306, 3'-CTACACC-5', 2430, 3'-CTAATTT-5', 2440, 3'-CCGCACC-5', 2566, 3'-TTATACC-5', 2590, 3'-CCACACC-5', 2602, 3'-CCACACT-5', 2636, 3'-TCAGATT-5', 2868, 3'-CTGCTCC-5', 2978, 3'-CCAGTCC-5', 2998, 3'-CCAGTCC-5', 3084, 3'-CTGGTCT-5', 3245, 3'-TCGCTCT-5', 3276, 3'-CTGGTCT-5', 3299, 3'-CTGCTCC-5', 3309, 3'-CTGCACC-5', 3322, 3'-CCGCATC-5', 3328, 3'-TTGCACT-5', 3343, 3'-CTGTTCC-5', 3352, 3'-TTGCATC-5', 3402, 3'-TCACACT-5', 3507, 3'-CCAGACC-5', 3550, 3'-CTGTTCC-5', 3625, 3'-TCACACC-5', 3824, 3'-TCATTTT-5', 4120, 3'-TCACTCT-5', 4128, 3'-TTGATTT-5', 4134, 3'-TTAGTTT-5', 4139, 3'-CTGCACC-5', 4343,
- positive strand in the negative direction is SuccessablesInr+-.bas, looking for 3'-C/T-C/T-A/G-A/C/G/T-A/T-C/T-C/T-5', 40, 3'-CTGAATT-5', 20, 3'-TTGGACC-5', 32, 3'-CTGCATT-5', 152, 3'-TTGAACC-5', 846, 3'-TCACACC-5', 882, 3'-TTGAACC-5', 1012, 3'-TCACTCC-5', 1058, 3'-TCACACC-5', 1128, 3'-TTGAACC-5', 1303, 3'-TTGCACC-5', 1339, 3'-TTGCACT-5', 1347, 3'-CCAGTCT-5', 1354, 3'-CCATTTC-5', 1380, 3'-TCGCTCT-5', 1450, 3'-CTATATC-5', 1528, 3'-TTATTTT-5', 1727, 3'-CTGCACT-5', 2000, 3'-CTACTCC-5', 2352, 3'-TTGAACC-5', 2382, 3'-TCACACC-5', 2418, 3'-CTGCACT-5', 2426, 3'-TTGAATC-5', 2708, 3'-TTGAACC-5', 2717, 3'-CTGCACC-5', 2761, 3'-TTGAACC-5', 3245, 3'-TTGCACT-5', 3289, 3'-CCAGATC-5', 3488, 3'-CTGCTCC-5', 3582, 3'-CCATTTC-5', 3688, 3'-CTGGACT-5', 3747, 3'-CTGAACC-5', 3784, 3'-CCATACC-5', 3858, 3'-TCACACC-5', 3967, 3'-CCGGACT-5', 4327, 3'-CTGCACT-5', 4340, 3'-CCAGTTC-5', 4417, 3'-CCACTCC-5', 4425, 3'-CCACTTT-5', 4461, 3'-TCACATT-5', 4533, 3'-TTAATTC-5', 4542,
- positive strand in the positive direction is SuccessablesInr++.bas, looking for 3'-C/T-C/T-A/G-A/C/G/T-A/T-C/T-C/T-5', 75, 3'-CTGGACC-5', 40, 3'-CCGGTCC-5', 215, 3'-TTACACT-5', 230, 3'-CCGGACC-5', 286, 3'-CCGTTCC-5', 503, 3'-TCGGTCC-5', 515, 3'-CCGCTCT-5', 557, 3'-CCGTTCC-5', 587, 3'-CCGCTCT-5', 641, 3'-CCGTTCC-5', 671, 3'-CCGGACT-5', 725, 3'-CCGTTCC-5', 823, 3'-TCGGTCT-5', 835, 3'-TTGGACC-5', 847, 3'-CCGTTCC-5', 923, 3'-TCGGTCT-5', 935, 3'-TTGGACC-5', 947, 3'-CCGTTCC-5', 1007, 3'-TCGCTCT-5', 1061, 3'-CCGGTCC-5', 1175, 3'-CCGCTCT-5', 1229, 3'-CCGTTCC-5', 1259, 3'-CCGTTCC-5', 1327, 3'-CCGCTCT-5', 1381, 3'-CCGTTCC-5', 1427, 3'-CCGCTCT-5', 1481, 3'-TCGTTCC-5', 1511, 3'-CCGCTCT-5', 1565, 3'-CCGCACT-5', 1720, 3'-CCACACC-5', 1805, 3'-CCGCTCT-5', 1921, 3'-CCGTTCT-5', 1948, 3'-CCACACC-5', 1971, 3'-TCAATTT-5', 2136, 3'-TTGTACT-5', 2141, 3'-CTACTTT-5', 2146, 3'-CCGTTCT-5', 2190, 3'-CCAGTCT-5', 2222, 3'-TTGGTCT-5', 2228, 3'-CCGCACT-5', 2555, 3'-CCGGTCC-5', 2574, 3'-TCAGTCT-5', 2609, 3'-TCAGTTC-5', 2615, 3'-TCAGTCC-5', 2620, 3'-CTATATT-5', 2662, 3'-TCAATCC-5', 2668, 3'-TCGTTTT-5', 2707, 3'-TCGATTC-5', 2789, 3'-TTGCTCC-5', 2806, 3'-CTAAACT-5', 2871, 3'-CTGGTCC-5', 2876, 3'-CCAGACT-5', 2943, 3'-CCGGACC-5', 2988, 3'-CCAGACC-5', 3021, 3'-TTATACC-5', 3162, 3'-CTGGTTT-5', 3175, 3'-TCGGTCT-5', 3221, 3'-CTACTCC-5', 3478, 3'-CCGATCC-5', 3484, 3'-TCGATCC-5', 3522, 3'-CTGGTCT-5', 3548, 3'-TCACACT-5', 3594, 3'-CCACTCC-5', 3647, 3'-CCGGACC-5', 3679, 3'-CCGGACC-5', 3758, 3'-CTGGACC-5', 3787, 3'-TCACTCC-5', 3878, 3'-TCAGACT-5', 3924, 3'-TCACACC-5', 3966, 3'-CCACACT-5', 3971, 3'-TTACTCC-5', 4096, 3'-CTACTCC-5', 4102, 3'-CTAAATC-5', 4136, 3'-CCACTCC-5', 4401, 3'-CCAGACC-5', 4416,
- complement, negative strand, negative direction is SuccessablesInrc--.bas, looking for 3'-A/G-A/G-C/T-A/C/G/T-A/T-A/G-A/G-5', 40, 3'-GACTTAA-5', 20, 3'-AACCTGG-5', 32, 3'-GACGTAA-5', 152, 3'-AACTTGG-5', 846, 3'-AGTGTGG-5', 882, 3'-AACTTGG-5', 1012, 3'-AGTGAGG-5', 1058, 3'-AGTGTGG-5', 1128, 3'-AACTTGG-5', 1303, 3'-AACGTGG-5', 1339, 3'-AACGTGA-5', 1347, 3'-GGTCAGA-5', 1354, 3'-GGTAAAG-5', 1380, 3'-AGCGAGA-5', 1450, 3'-GATATAG-5', 1528, 3'-AATAAAA-5', 1727, 3'-GACGTGA-5', 2000, 3'-GATGAGG-5', 2352, 3'-AACTTGG-5', 2382, 3'-AGTGTGG-5', 2418, 3'-GACGTGA-5', 2426, 3'-AACTTAG-5', 2708, 3'-AACTTGG-5', 2717, 3'-GACGTGG-5', 2761, 3'-AACTTGG-5', 3245, 3'-AACGTGA-5', 3289, 3'-GGTCTAG-5', 3488, 3'-GACGAGG-5', 3582, 3'-GGTAAAG-5', 3688, 3'-GACCTGA-5', 3747, 3'-GACTTGG-5', 3784, 3'-GGTATGG-5', 3858, 3'-AGTGTGG-5', 3967, 3'-GGCCTGA-5', 4327, 3'-GACGTGA-5', 4340, 3'-GGTCAAG-5', 4417, 3'-GGTGAGG-5', 4425, 3'-GGTGAAA-5', 4461, 3'-AGTGTAA-5', 4533, 3'-AATTAAG-5', 4542,
- complement, negative strand, positive direction is SuccessablesInrc-+.bas, looking for 3'-A/G-A/G-C/T-A/C/G/T-A/T-A/G-A/G-5', 75, 3'-GACCTGG-5', 40, 3'-GGCCAGG-5', 215, 3'-AATGTGA-5', 230, 3'-GGCCTGG-5', 286, 3'-GGCAAGG-5', 503, 3'-AGCCAGG-5', 515, 3'-GGCGAGA-5', 557, 3'-GGCAAGG-5', 587, 3'-GGCGAGA-5', 641, 3'-GGCAAGG-5', 671, 3'-GGCCTGA-5', 725, 3'-GGCAAGG-5', 823, 3'-AGCCAGA-5', 835, 3'-AACCTGG-5', 847, 3'-GGCAAGG-5', 923, 3'-AGCCAGA-5', 935, 3'-AACCTGG-5', 947, 3'-GGCAAGG-5', 1007, 3'-AGCGAGA-5', 1061, 3'-GGCCAGG-5', 1175, 3'-GGCGAGA-5', 1229, 3'-GGCAAGG-5', 1259, 3'-GGCAAGG-5', 1327, 3'-GGCGAGA-5', 1381, 3'-GGCAAGG-5', 1427, 3'-GGCGAGA-5', 1481, 3'-AGCAAGG-5', 1511, 3'-GGCGAGA-5', 1565, 3'-GGCGTGA-5', 1720, 3'-GGTGTGG-5', 1805, 3'-GGCGAGA-5', 1921, 3'-GGCAAGA-5', 1948, 3'-GGTGTGG-5', 1971, 3'-AGTTAAA-5', 2136, 3'-AACATGA-5', 2141, 3'-GATGAAA-5', 2146, 3'-GGCAAGA-5', 2190, 3'-GGTCAGA-5', 2222, 3'-AACCAGA-5', 2228, 3'-GGCGTGA-5', 2555, 3'-GGCCAGG-5', 2574, 3'-AGTCAGA-5', 2609, 3'-AGTCAAG-5', 2615, 3'-AGTCAGG-5', 2620, 3'-GATATAA-5', 2662, 3'-AGTTAGG-5', 2668, 3'-AGCAAAA-5', 2707, 3'-AGCTAAG-5', 2789, 3'-AACGAGG-5', 2806, 3'-GATTTGA-5', 2871, 3'-GACCAGG-5', 2876, 3'-GGTCTGA-5', 2943, 3'-GGCCTGG-5', 2988, 3'-GGTCTGG-5', 3021, 3'-AATATGG-5', 3162, 3'-GACCAAA-5', 3175, 3'-AGCCAGA-5', 3221, 3'-GATGAGG-5', 3478, 3'-GGCTAGG-5', 3484, 3'-AGCTAGG-5', 3522, 3'-GACCAGA-5', 3548, 3'-AGTGTGA-5', 3594, 3'-GGTGAGG-5', 3647, 3'-GGCCTGG-5', 3679, 3'-GGCCTGG-5', 3758, 3'-GACCTGG-5', 3787, 3'-AGTGAGG-5', 3878, 3'-AGTCTGA-5', 3924, 3'-AGTGTGG-5', 3966, 3'-GGTGTGA-5', 3971, 3'-AATGAGG-5', 4096, 3'-GATGAGG-5', 4102, 3'-GATTTAG-5', 4136, 3'-GGTGAGG-5', 4401, 3'-GGTCTGG-5', 4416,
- complement, positive strand, negative direction is SuccessablesInrc+-.bas, looking for 3'-A/G-A/G-C/T-A/C/G/T-A/T-A/G-A/G-5', 121, 3'-AACAAGG-5', 71, 3'-GATATGG-5', 77, 3'-GGCAAAG-5', 93, 3'-GGCATGA-5', 124, 3'-GGTATAA-5', 181, 3'-GATGTAA-5', 247, 3'-AACCAGG-5', 262, 3'-AATATGA-5', 274, 3'-AGTGAGA-5', 301, 3'-GACGAAA-5', 312, 3'-GGCCAAG-5', 419, 3'-GGTCAGG-5', 441, 3'-AGCCTGG-5', 459, 3'-AACATAG-5', 468, 3'-AGTGAAA-5', 473, 3'-AGCCTGG-5', 508, 3'-GGCCAAG-5', 556, 3'-GGTCAGG-5', 578, 3'-AATATGG-5', 605, 3'-GGCCAGG-5', 648, 3'-GGCCAAG-5', 692, 3'-GGTCAGG-5', 714, 3'-AGCCTGA-5', 732, 3'-AGCGTGG-5', 741, 3'-GATGTGG-5', 787, 3'-AGCCAAG-5', 874, 3'-AGCCTGG-5', 899, 3'-AGCGAGA-5', 913, 3'-AGCCAGG-5', 948, 3'-GGCATGG-5', 953, 3'-AATCAGG-5', 984, 3'-AACCTGG-5', 1015, 3'-AGTGAGA-5', 1079, 3'-AGCCTGG-5', 1198, 3'-AACATGG-5', 1207, 3'-GGTGAAA-5', 1212, 3'-GGCGTGG-5', 1244, 3'-AACCTAG-5', 1306, 3'-AGTCTGG-5', 1356, 3'-AATAAGA-5', 1365, 3'-AGCAAAA-5', 1371, 3'-AACAAAA-5', 1394, 3'-GGTGTGA-5', 1479, 3'-AACGAAG-5', 1555, 3'-GGCAAAA-5', 1561, 3'-AATGAAA-5', 1582, 3'-AACCTAA-5', 1591, 3'-AATTAAA-5', 1697, 3'-AATATGG-5', 1742, 3'-GGCGTGG-5', 1897, 3'-GGCATGA-5', 1953, 3'-AACCTGG-5', 1959, 3'-AGCCTGG-5', 2009, 3'-AGCAAGA-5', 2023, 3'-AATGTGG-5', 2065, 3'-GGCCAGG-5', 2077, 3'-AGTGTAA-5', 2087, 3'-AGTTTGA-5', 2141, 3'-AACATGG-5', 2152, 3'-GGCGAAA-5', 2157, 3'-GGTCAGG-5', 2250, 3'-AGTTTGA-5', 2257, 3'-AGCCTGG-5', 2268, 3'-AGCATGG-5', 2277, 3'-GGTGAAA-5', 2282, 3'-AACCTGG-5', 2385, 3'-AGCCTGG-5', 2435, 3'-AGTGAGA-5', 2449, 3'-AGCAAAA-5', 2476, 3'-AACAAAA-5', 2490, 3'-AGTAAGA-5', 2503, 3'-GGCCAGG-5', 2519, 3'-GGTCAGG-5', 2587, 3'-AGTGTGG-5', 2605, 3'-AACATGG-5', 2614, 3'-GGTGAAA-5', 2619, 3'-AGTGTGG-5', 2658, 3'-AACCTGG-5', 2720, 3'-AGCCTGG-5', 2770, 3'-AGCATGA-5', 2784, 3'-AACTAAG-5', 2914, 3'-GGCTAAA-5', 3009, 3'-AACTAAG-5', 3031, 3'-GGCGTGG-5', 3047, 3'-AGCCTGG-5', 3128, 3'-AACAAGG-5', 3141, 3'-GGTGAAA-5', 3146, 3'-AACATAA-5', 3169, 3'-GGTGTGG-5', 3186, 3'-AGCCAAG-5', 3273, 3'-AGCCTGG-5', 3298, 3'-AACAAGA-5', 3307, 3'-AGCAAAA-5', 3313, 3'-AACAAGA-5', 3340, 3'-AGCAAGA-5', 3374, 3'-GGCTTGA-5', 3401, 3'-GGCATAG-5', 3446, 3'-AACTAGA-5', 3463, 3'-AACCAGA-5', 3486, 3'-GACAAGA-5', 3759, 3'-GATGTGG-5', 3810, 3'-GACCAGG-5', 3871, 3'-AGTAAGA-5', 3893, 3'-GATGAAA-5', 3922, 3'-GGCCAGG-5', 3951, 3'-AGCCTGG-5', 4037, 3'-AACATAG-5', 4046, 3'-AGTGAGA-5', 4051, 3'-AATGTGA-5', 4092, 3'-GGCCAGG-5', 4102, 3'-GGCATGG-5', 4107, 3'-GGCCAGG-5', 4170, 3'-AGCTTGG-5', 4188, 3'-AGTGAGA-5', 4202, 3'-AGCCAGA-5', 4233, 3'-GACGTGG-5', 4238, 3'-AGCCTGG-5', 4300, 3'-GGTCAAA-5', 4309, 3'-AGCCTGG-5', 4349, 3'-AGTGTGA-5', 4361, 3'-AATGAGG-5', 4557,
- complement, positive strand, positive direction is SuccessablesInrc++.bas, looking for 3'-A/G-A/G-C/T-A/C/G/T-A/T-A/G-A/G-5', 45, 3'-AACATAA-5', 115, 3'-GACAAAA-5', 147, 3'-GGTGTGA-5', 345, 3'-GGCCTGA-5', 746, 3'-GACGTGA-5', 1372, 3'-GACGTGA-5', 1472, 3'-GGTCTGA-5', 1744, 3'-GGTGAAG-5', 1914, 3'-GATAAAG-5', 1978, 3'-GGTCAGG-5', 2026, 3'-AGCGAAG-5', 2095, 3'-AGTATAA-5', 2178, 3'-GACGTAA-5', 2206, 3'-GGTCTAG-5', 2230, 3'-AGTTAGA-5', 2235, 3'-GACAAAG-5', 2263, 3'-AGTGAGA-5', 2306, 3'-GATGTGG-5', 2430, 3'-GATTAAA-5', 2440, 3'-GGCGTGG-5', 2566, 3'-AATATGG-5', 2590, 3'-GGTGTGG-5', 2602, 3'-GGTGTGA-5', 2636, 3'-AGTCTAA-5', 2868, 3'-GACGAGG-5', 2978, 3'-GGTCAGG-5', 2998, 3'-GGTCAGG-5', 3084, 3'-GACCAGA-5', 3245, 3'-AGCGAGA-5', 3276, 3'-GACCAGA-5', 3299, 3'-GACGAGG-5', 3309, 3'-GACGTGG-5', 3322, 3'-GGCGTAG-5', 3328, 3'-AACGTGA-5', 3343, 3'-GACAAGG-5', 3352, 3'-AACGTAG-5', 3402, 3'-AGTGTGA-5', 3507, 3'-GGTCTGG-5', 3550, 3'-GACAAGG-5', 3625, 3'-AGTGTGG-5', 3824, 3'-AGTAAAA-5', 4120, 3'-AGTGAGA-5', 4128, 3'-AACTAAA-5', 4134, 3'-AATCAAA-5', 4139, 3'-GACGTGG-5', 4343,
- inverse complement, negative strand, negative direction is SuccessablesInrci--.bas, looking for 3'-A/G-A/G-A/T-A/C/G/T-C/T-A/G-A/G-5', 32, 3'-GATACAA-5', 213, 3'-GGACCGA-5', 598, 3'-AGTGCGG-5', 664, 3'-GGACTGG-5', 734, 3'-AGTGTGG-5', 882, 3'-GAAGTGA-5', 1056, 3'-AGTGTGG-5', 1128, 3'-GGACCGG-5', 1200, 3'-AGAGCGA-5', 1448, 3'-GGTCCGA-5', 1462, 3'-GATATAG-5', 1528, 3'-AGAACGG-5', 1608, 3'-AAAATAG-5', 1730, 3'-AGTGCAG-5', 1773, 3'-GGACCGA-5', 1843, 3'-AGTGCGG-5', 1992, 3'-AGTGCGG-5', 2208, 3'-AGTGTGG-5', 2418, 3'-AGTACGG-5', 2535, 3'-AGTACGG-5', 2753, 3'-AAAGTAG-5', 2887, 3'-GATTCGA-5', 3033, 3'-GGACCGG-5', 3130, 3'-AGTGCGG-5', 3281, 3'-AGTCCGA-5', 3398, 3'-GGTCTAG-5', 3488, 3'-GGTATGG-5', 3858, 3'-GGTCCGG-5', 3873, 3'-AGTGTGG-5', 3967, 3'-AGTACGG-5', 4118, 3'-GGTCCGA-5', 4255, 3'-AGTGTAA-5', 4533,
- inverse complement, negative strand, positive direction is SuccessablesInrci-+.bas, looking for 3'-A/G-A/G-A/T-A/C/G/T-C/T-A/G-A/G-5', 61, 3'-AGAGTGG-5', 53, 3'-AATGTGA-5', 230, 3'-GGAGCGA-5', 429, 3'-AGACCGG-5', 442, 3'-GGTGCGG-5', 489, 3'-AGTGCGG-5', 498, 3'-AGTGCGG-5', 582, 3'-AGTGCGG-5', 666, 3'-GGTGCAG-5', 784, 3'-AGTGCGG-5', 1086, 3'-AGTGCGG-5', 1170, 3'-AGTGCGG-5', 1254, 3'-AATGCGG-5', 1322, 3'-AATGCGG-5', 1422, 3'-AGTGCGG-5', 1590, 3'-GAAGCGG-5', 1636, 3'-GGTGCGG-5', 1764, 3'-AGTGCAG-5', 1787, 3'-GGTGTGG-5', 1805, 3'-GAACTGG-5', 1953, 3'-GGTGTGG-5', 1971, 3'-AAAGCAG-5', 2007, 3'-AGTGCAG-5', 2064, 3'-GAACCAG-5', 2227, 3'-AGATCAA-5', 2232, 3'-AGTGCAG-5', 2327, 3'-GGTGCAA-5', 2335, 3'-GAAATAG-5', 2626, 3'-GATATAA-5', 2662, 3'-GGACTGA-5', 2674, 3'-AGAGCAA-5', 2705, 3'-AAAGTGG-5', 2711, 3'-GGTGCAA-5', 2801, 3'-AGAATGA-5', 2841, 3'-GATTTGA-5', 2871, 3'-GGTCTGA-5', 2943, 3'-GGTCTGG-5', 3021, 3'-AATATGG-5', 3162, 3'-GAAATGG-5', 3168, 3'-GGACCAA-5', 3174, 3'-GGAATGA-5', 3441, 3'-GATGCAG-5', 3460, 3'-AGTGCAG-5', 3465, 3'-GGACCAG-5', 3547, 3'-GGAATGA-5', 3567, 3'-AGTGTGA-5', 3594, 3'-GAAGCGG-5', 3670, 3'-AATCCGA-5', 3799, 3'-AGAATGA-5', 3835, 3'-GAACCAG-5', 3840, 3'-AGAGTGA-5', 3876, 3'-AGTCTGA-5', 3924, 3'-AGTGTGG-5', 3966, 3'-GGTGTGA-5', 3971, 3'-AGAGTGG-5', 4040, 3'-AGAACAG-5', 4069, 3'-GAAATGA-5', 4094, 3'-GATTTAG-5', 4136, 3'-GGAGTGA-5', 4350, 3'-GGTCTGG-5', 4416, 3'-GGAACAA-5', 4445,
- inverse complement, positive strand, negative direction is SuccessablesInrci+-.bas, looking for 3'-A/G-A/G-A/T-A/C/G/T-C/T-A/G-A/G-5', 100, 3'-AGACTGA-5', 17, 3'-GGACCAG-5', 34, 3'-AAAACAA-5', 69, 3'-GATATGG-5', 77, 3'-AAACTGA-5', 130, 3'-AAAACAG-5', 167, 3'-GGTATAA-5', 181, 3'-GAAACAA-5', 229, 3'-GATGTAA-5', 247, 3'-AGTTCAA-5', 255, 3'-AAACCAG-5', 261, 3'-AATATGA-5', 274, 3'-AGAACAG-5', 288, 3'-AAACTGA-5', 307, 3'-GGTGCGG-5', 380, 3'-AGTGCGA-5', 448, 3'-AATACGA-5', 492, 3'-AAATTAG-5', 499, 3'-AGATTGA-5', 585, 3'-AATATGG-5', 605, 3'-AATACAA-5', 635, 3'-AAATTGG-5', 643, 3'-AGTTCGA-5', 721, 3'-AGACCAG-5', 727, 3'-AATACAA-5', 769, 3'-AAATTAG-5', 777, 3'-GATGTGG-5', 787, 3'-AGAGCGA-5', 911, 3'-GATCCAG-5', 975, 3'-AGATTGG-5', 1045, 3'-AGAGTGA-5', 1077, 3'-AAATTAG-5', 1234, 3'-AGTCTGG-5', 1356, 3'-AGAGCAA-5', 1369, 3'-AAAACAA-5', 1388, 3'-AGTGCAG-5', 1471, 3'-GGTGTGA-5', 1479, 3'-AGTGCAA-5', 1536, 3'-AGAACGA-5', 1553, 3'-AATACAG-5', 1566, 3'-GAAACAA-5', 1585, 3'-GAAATGA-5', 1663, 3'-AAAGCGG-5', 1680, 3'-GAATTAA-5', 1696, 3'-AATATGG-5', 1742, 3'-AATACAA-5', 1878, 3'-AAATTAG-5', 1887, 3'-AGACTGA-5', 1935, 3'-AGAATGG-5', 1948, 3'-AGAGCAA-5', 2021, 3'-AATGTGG-5', 2065, 3'-GGTGCAG-5', 2082, 3'-AGTGTAA-5', 2087, 3'-AGTTTGA-5', 2141, 3'-AGACCAA-5', 2147, 3'-GATACAA-5', 2180, 3'-AAAATGA-5', 2187, 3'-GGTGCGG-5', 2197, 3'-AGTTTGA-5', 2257, 3'-AGACCAG-5', 2263, 3'-AATACAA-5', 2305, 3'-AAACTAG-5', 2313, 3'-AGAGTGA-5', 2447, 3'-GATTCGG-5', 2454, 3'-AAAGCAA-5', 2474, 3'-AAAGCAA-5', 2480, 3'-AAAACAA-5', 2509, 3'-AGACCAG-5', 2600, 3'-AGTGTGG-5', 2605, 3'-AAATCAG-5', 2649, 3'-AGTGTGG-5', 2658, 3'-AAAACAA-5', 2842, 3'-AGAATGG-5', 3004, 3'-AAAATAA-5', 3013, 3'-AAACTAA-5', 3030, 3'-AGACCAG-5', 3123, 3'-AAATTAG-5', 3176, 3'-GGTGTGG-5', 3186, 3'-AGAGCAA-5', 3311, 3'-AAAACAA-5', 3330, 3'-AAATTGA-5', 3358, 3'-GAAGTGA-5', 3410, 3'-GAACTAG-5', 3462, 3'-AAACCAG-5', 3485, 3'-AATCCAG-5', 3681, 3'-GGAACAG-5', 3725, 3'-GGACTGG-5', 3749, 3'-AATGCAG-5', 3772, 3'-GATGTGG-5', 3810, 3'-GGACCAG-5', 3870, 3'-GGAGTAA-5', 3891, 3'-AGTTCAA-5', 4026, 3'-AGACCAG-5', 4032, 3'-AAAATAA-5', 4071, 3'-AATGTGA-5', 4092, 3'-AGTTCAA-5', 4177, 3'-AAAATAA-5', 4221, 3'-AGTGTGA-5', 4361, 3'-AGTCCAA-5', 4502, 3'-GGAATGA-5', 4555,
- inverse complement, positive strand, positive direction is SuccessablesInrci++.bas, looking for 3'-A/G-A/G-A/T-A/C/G/T-C/T-A/G-A/G-5', 75, 3'-GGTCCGA-5', 10, 3'-AGTCCGG-5', 92, 3'-AATCCAG-5', 152, 3'-GGTCCAG-5', 217, 3'-GGTGTGA-5', 345, 3'-GAAGCGG-5', 459, 3'-AGAATGA-5', 524, 3'-GAAGCGG-5', 595, 3'-GATGCGA-5', 652, 3'-GGTGCGA-5', 777, 3'-GGACCGG-5', 849, 3'-GGACCGG-5', 949, 3'-GGTCCGA-5', 1177, 3'-AAAGCAG-5', 1183, 3'-GAAGCGG-5', 1308, 3'-GAAGCGG-5', 1408, 3'-AATTCGG-5', 1541, 3'-GATGCGA-5', 1576, 3'-GGACTGG-5', 1662, 3'-GGTCTGA-5', 1744, 3'-GGACCGA-5', 1817, 3'-GGTCCGG-5', 1857, 3'-AGAATGG-5', 1888, 3'-GAAGTAG-5', 2110, 3'-AGTATAA-5', 2178, 3'-GGACTGG-5', 2213, 3'-GGTCTAG-5', 2230, 3'-AGAGTGG-5', 2247, 3'-AAAGTGA-5', 2304, 3'-GGTCCGA-5', 2318, 3'-AATCCGA-5', 2368, 3'-GATGTGG-5', 2430, 3'-GGACCGA-5', 2435, 3'-AGAGTGG-5', 2470, 3'-GGTACAA-5', 2475, 3'-GGACCGG-5', 2571, 3'-AATATGG-5', 2590, 3'-GGTGTGG-5', 2602, 3'-AGTTCAG-5', 2617, 3'-GGTGTGA-5', 2636, 3'-AGTCTAA-5', 2868, 3'-AAACTGG-5', 2873, 3'-GGTCCGG-5', 2878, 3'-AGACCGA-5', 2885, 3'-GGAGTAA-5', 2902, 3'-AGACTGA-5', 2945, 3'-AGACCGG-5', 2985, 3'-GGACCGG-5', 2990, 3'-GGAACAG-5', 3003, 3'-GGTCCAG-5', 3018, 3'-AGACCAA-5', 3023, 3'-AGTCCGG-5', 3036, 3'-GGACCAA-5', 3049, 3'-GAAGTAG-5', 3250, 3'-AGTGCAG-5', 3255, 3'-GGACCAG-5', 3298, 3'-AGAGTGA-5', 3317, 3'-GGTACAA-5', 3337, 3'-GGAACGG-5', 3375, 3'-AGTGTGA-5', 3507, 3'-GATCCGA-5', 3524, 3'-GGTCTGG-5', 3550, 3'-AGAGTGG-5', 3612, 3'-GGACCGG-5', 3681, 3'-AGTGTGG-5', 3824, 3'-GAACTGG-5', 4018, 3'-AAAATAG-5', 4123, 3'-GAACTAA-5', 4133, 3'-AAATCAA-5', 4138, 3'-GAAACGG-5', 4210, 3'-GGACTGG-5', 4216, 3'-GGAGTAA-5', 4309, 3'-AGTACAG-5', 4366, 3'-GGTACGA-5', 4372, 3'-AGAACGA-5', 4390,
- inverse, negative strand, negative direction, is SuccessablesInri--.bas, looking for 3'-C/T-C/T-A/T-A/C/G/T-A/G-C/T-C/T-5', 100, 3'-TCTGACT-5', 17, 3'-CCTGGTC-5', 34, 3'-TTTTGTT-5', 69, 3'-CTATACC-5', 77, 3'-TTTGACT-5', 130, 3'-TTTTGTC-5', 167, 3'-CCATATT-5', 181, 3'-CTTTGTT-5', 229, 3'-CTACATT-5', 247, 3'-TCAAGTT-5', 255, 3'-TTTGGTC-5', 261, 3'-TTATACT-5', 274, 3'-TCTTGTC-5', 288, 3'-TTTGACT-5', 307, 3'-CCACGCC-5', 380, 3'-TCACGCT-5', 448, 3'-TTATGCT-5', 492, 3'-TTTAATC-5', 499, 3'-TCTAACT-5', 585, 3'-TTATACC-5', 605, 3'-TTATGTT-5', 635, 3'-TTTAACC-5', 643, 3'-TCAAGCT-5', 721, 3'-TCTGGTC-5', 727, 3'-TTATGTT-5', 769, 3'-TTTAATC-5', 777, 3'-CTACACC-5', 787, 3'-TCTCGCT-5', 911, 3'-CTAGGTC-5', 975, 3'-TCTAACC-5', 1045, 3'-TCTCACT-5', 1077, 3'-TTTAATC-5', 1234, 3'-TCAGACC-5', 1356, 3'-TCTCGTT-5', 1369, 3'-TTTTGTT-5', 1388, 3'-TCACGTC-5', 1471, 3'-CCACACT-5', 1479, 3'-TCACGTT-5', 1536, 3'-TCTTGCT-5', 1553, 3'-TTATGTC-5', 1566, 3'-CTTTGTT-5', 1585, 3'-CTTTACT-5', 1663, 3'-TTTCGCC-5', 1680, 3'-CTTAATT-5', 1696, 3'-TTATACC-5', 1742, 3'-TTATGTT-5', 1878, 3'-TTTAATC-5', 1887, 3'-TCTGACT-5', 1935, 3'-TCTTACC-5', 1948, 3'-TCTCGTT-5', 2021, 3'-TTACACC-5', 2065, 3'-CCACGTC-5', 2082, 3'-TCACATT-5', 2087, 3'-TCAAACT-5', 2141, 3'-TCTGGTT-5', 2147, 3'-CTATGTT-5', 2180, 3'-TTTTACT-5', 2187, 3'-CCACGCC-5', 2197, 3'-TCAAACT-5', 2257, 3'-TCTGGTC-5', 2263, 3'-TTATGTT-5', 2305, 3'-TTTGATC-5', 2313, 3'-TCTCACT-5', 2447, 3'-CTAAGCC-5', 2454, 3'-TTTCGTT-5', 2474, 3'-TTTCGTT-5', 2480, 3'-TTTTGTT-5', 2509, 3'-TCTGGTC-5', 2600, 3'-TCACACC-5', 2605, 3'-TTTAGTC-5', 2649, 3'-TCACACC-5', 2658, 3'-TTTTGTT-5', 2842, 3'-TCTTACC-5', 3004, 3'-TTTTATT-5', 3013, 3'-TTTGATT-5', 3030, 3'-TCTGGTC-5', 3123, 3'-TTTAATC-5', 3176, 3'-CCACACC-5', 3186, 3'-TCTCGTT-5', 3311, 3'-TTTTGTT-5', 3330, 3'-TTTAACT-5', 3358, 3'-CTTCACT-5', 3410, 3'-CTTGATC-5', 3462, 3'-TTTGGTC-5', 3485, 3'-TTAGGTC-5', 3681, 3'-CCTTGTC-5', 3725, 3'-CCTGACC-5', 3749, 3'-TTACGTC-5', 3772, 3'-CTACACC-5', 3810, 3'-CCTGGTC-5', 3870, 3'-CCTCATT-5', 3891, 3'-TCAAGTT-5', 4026, 3'-TCTGGTC-5', 4032, 3'-TTTTATT-5', 4071, 3'-TTACACT-5', 4092, 3'-TCAAGTT-5', 4177, 3'-TTTTATT-5', 4221, 3'-TCACACT-5', 4361, 3'-TCAGGTT-5', 4502, 3'-CCTTACT-5', 4555,
- inverse, negative strand, positive direction, is SuccessablesInri-+.bas, looking for 3'-C/T-C/T-A/T-A/C/G/T-A/G-C/T-C/T-5', 75, 3'-CCAGGCT-5', 10, 3'-TCAGGCC-5', 92, 3'-TTAGGTC-5', 152, 3'-CCAGGTC-5', 217, 3'-CCACACT-5', 345, 3'-CTTCGCC-5', 459, 3'-TCTTACT-5', 524, 3'-CTTCGCC-5', 595, 3'-CTACGCT-5', 652, 3'-CCACGCT-5', 777, 3'-CCTGGCC-5', 849, 3'-CCTGGCC-5', 949, 3'-CCAGGCT-5', 1177, 3'-TTTCGTC-5', 1183, 3'-CTTCGCC-5', 1308, 3'-CTTCGCC-5', 1408, 3'-TTAAGCC-5', 1541, 3'-CTACGCT-5', 1576, 3'-CCTGACC-5', 1662, 3'-CCAGACT-5', 1744, 3'-CCTGGCT-5', 1817, 3'-CCAGGCC-5', 1857, 3'-TCTTACC-5', 1888, 3'-CTTCATC-5', 2110, 3'-TCATATT-5', 2178, 3'-CCTGACC-5', 2213, 3'-CCAGATC-5', 2230, 3'-TCTCACC-5', 2247, 3'-TTTCACT-5', 2304, 3'-CCAGGCT-5', 2318, 3'-TTAGGCT-5', 2368, 3'-CTACACC-5', 2430, 3'-CCTGGCT-5', 2435, 3'-TCTCACC-5', 2470, 3'-CCATGTT-5', 2475, 3'-CCTGGCC-5', 2571, 3'-TTATACC-5', 2590, 3'-CCACACC-5', 2602, 3'-TCAAGTC-5', 2617, 3'-CCACACT-5', 2636, 3'-TCAGATT-5', 2868, 3'-TTTGACC-5', 2873, 3'-CCAGGCC-5', 2878, 3'-TCTGGCT-5', 2885, 3'-CCTCATT-5', 2902, 3'-TCTGACT-5', 2945, 3'-TCTGGCC-5', 2985, 3'-CCTGGCC-5', 2990, 3'-CCTTGTC-5', 3003, 3'-CCAGGTC-5', 3018, 3'-TCTGGTT-5', 3023, 3'-TCAGGCC-5', 3036, 3'-CCTGGTT-5', 3049, 3'-CTTCATC-5', 3250, 3'-TCACGTC-5', 3255, 3'-CCTGGTC-5', 3298, 3'-TCTCACT-5', 3317, 3'-CCATGTT-5', 3337, 3'-CCTTGCC-5', 3375, 3'-TCACACT-5', 3507, 3'-CTAGGCT-5', 3524, 3'-CCAGACC-5', 3550, 3'-TCTCACC-5', 3612, 3'-CCTGGCC-5', 3681, 3'-TCACACC-5', 3824, 3'-CTTGACC-5', 4018, 3'-TTTTATC-5', 4123, 3'-CTTGATT-5', 4133, 3'-TTTAGTT-5', 4138, 3'-CTTTGCC-5', 4210, 3'-CCTGACC-5', 4216, 3'-CCTCATT-5', 4309, 3'-TCATGTC-5', 4366, 3'-CCATGCT-5', 4372, 3'-TCTTGCT-5', 4390,
- inverse, positive strand, negative direction, is SuccessablesInri+-.bas, looking for 3'-C/T-C/T-A/T-A/C/G/T-A/G-C/T-C/T-5', 32, 3'-CTATGTT-5', 213, 3'-CCTGGCT-5', 598, 3'-TCACGCC-5', 664, 3'-CCTGACC-5', 734, 3'-TCACACC-5', 882, 3'-CTTCACT-5', 1056, 3'-TCACACC-5', 1128, 3'-CCTGGCC-5', 1200, 3'-TCTCGCT-5', 1448, 3'-CCAGGCT-5', 1462, 3'-CTATATC-5', 1528, 3'-TCTTGCC-5', 1608, 3'-TTTTATC-5', 1730, 3'-TCACGTC-5', 1773, 3'-CCTGGCT-5', 1843, 3'-TCACGCC-5', 1992, 3'-TCACGCC-5', 2208, 3'-TCACACC-5', 2418, 3'-TCATGCC-5', 2535, 3'-TCATGCC-5', 2753, 3'-TTTCATC-5', 2887, 3'-CTAAGCT-5', 3033, 3'-CCTGGCC-5', 3130, 3'-TCACGCC-5', 3281, 3'-TCAGGCT-5', 3398, 3'-CCAGATC-5', 3488, 3'-CCATACC-5', 3858, 3'-CCAGGCC-5', 3873, 3'-TCACACC-5', 3967, 3'-TCATGCC-5', 4118, 3'-CCAGGCT-5', 4255, 3'-TCACATT-5', 4533,
- inverse, positive strand, positive direction, is SuccessablesInri++.bas, looking for 3'-C/T-C/T-A/T-A/C/G/T-A/G-C/T-C/T-5', 61, 3'-TCTCACC-5', 53, 3'-TTACACT-5', 230, 3'-CCTCGCT-5', 429, 3'-TCTGGCC-5', 442, 3'-CCACGCC-5', 489, 3'-TCACGCC-5', 498, 3'-TCACGCC-5', 582, 3'-TCACGCC-5', 666, 3'-CCACGTC-5', 784, 3'-TCACGCC-5', 1086, 3'-TCACGCC-5', 1170, 3'-TCACGCC-5', 1254, 3'-TTACGCC-5', 1322, 3'-TTACGCC-5', 1422, 3'-TCACGCC-5', 1590, 3'-CTTCGCC-5', 1636, 3'-CCACGCC-5', 1764, 3'-TCACGTC-5', 1787, 3'-CCACACC-5', 1805, 3'-CTTGACC-5', 1953, 3'-CCACACC-5', 1971, 3'-TTTCGTC-5', 2007, 3'-TCACGTC-5', 2064, 3'-CTTGGTC-5', 2227, 3'-TCTAGTT-5', 2232, 3'-TCACGTC-5', 2327, 3'-CCACGTT-5', 2335, 3'-CTTTATC-5', 2626, 3'-CTATATT-5', 2662, 3'-CCTGACT-5', 2674, 3'-TCTCGTT-5', 2705, 3'-TTTCACC-5', 2711, 3'-CCACGTT-5', 2801, 3'-TCTTACT-5', 2841, 3'-CTAAACT-5', 2871, 3'-CCAGACT-5', 2943, 3'-CCAGACC-5', 3021, 3'-TTATACC-5', 3162, 3'-CTTTACC-5', 3168, 3'-CCTGGTT-5', 3174, 3'-CCTTACT-5', 3441, 3'-CTACGTC-5', 3460, 3'-TCACGTC-5', 3465, 3'-CCTGGTC-5', 3547, 3'-CCTTACT-5', 3567, 3'-TCACACT-5', 3594, 3'-CTTCGCC-5', 3670, 3'-TTAGGCT-5', 3799, 3'-TCTTACT-5', 3835, 3'-CTTGGTC-5', 3840, 3'-TCTCACT-5', 3876, 3'-TCAGACT-5', 3924, 3'-TCACACC-5', 3966, 3'-CCACACT-5', 3971, 3'-TCTCACC-5', 4040, 3'-TCTTGTC-5', 4069, 3'-CTTTACT-5', 4094, 3'-CTAAATC-5', 4136, 3'-CCTCACT-5', 4350, 3'-CCAGACC-5', 4416, 3'-CCTTGTT-5', 4445.
For the Basic programs testing consensus sequence BBCABW (starting with SuccessablesInr2.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesInr2--.bas, looking for 3'-C/G/T-C/G/T-C-A-C/G/T-A/T-5', 44, 3'-TCCATA-5', 179, 3'-CCCAGT-5', 206, 3'-CTCAGA-5', 278, 3'-GTCACT-5', 299, 3'-TTCACA-5', 322, 3'-TCCAGT-5', 439, 3'-TGCATT-5', 533, 3'-TCCAGT-5', 568, 3'-TCCAGT-5', 576, 3'-TCCAGT-5', 712, 3'-GGCAGA-5', 754, 3'-GCCACT-5', 868, 3'-GTCACT-5', 1034, 3'-CCCACT-5', 1049, 3'-CTCACT-5', 1077, 3'-GGCACA-5', 1220, 3'-GTCACT-5', 1325, 3'-GTCAGA-5', 1354, 3'-CTCAGA-5', 1444, 3'-GGCAGT-5', 1511, 3'-TGCAGA-5', 1774, 3'-GTCACT-5', 1978, 3'-GTCACA-5', 2085, 3'-TCCAGT-5', 2248, 3'-GTCACT-5', 2404, 3'-CTCACT-5', 2447, 3'-TCCAGT-5', 2585, 3'-GTCACA-5', 2603, 3'-GTCACA-5', 2656, 3'-GTCACT-5', 2739, 3'-TTCACA-5', 2860, 3'-TCCACT-5', 3144, 3'-CCCACA-5', 3184, 3'-TTCACT-5', 3410, 3'-GTCATT-5', 3480, 3'-TCCACT-5', 3825, 3'-CTCATA-5', 3829, 3'-CTCATT-5', 3891, 3'-TTCACA-5', 3939, 3'-GTCACT-5', 4200, 3'-TCCAGT-5', 4307, 3'-GTCACT-5', 4319, 3'-CCCACT-5', 4353, 3'-GTCACA-5', 4359,
- negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesInr2-+.bas, looking for 3'-C/G/T-C/G/T-C-A-C/G/T-A/T-5', 87, 3'-TCCAGA-5', 15, 3'-GGCATT-5', 22, 3'-GTCACA-5', 155, 3'-CCCAGA-5', 204, 3'-GCCACA-5', 343, 3'-CGCAGA-5', 396, 3'-TGCAGA-5', 438, 3'-CCCAGA-5', 468, 3'-TGCACA-5', 548, 3'-TCCACA-5', 632, 3'-CGCACT-5', 686, 3'-CGCACA-5', 800, 3'-GCCAGA-5', 835, 3'-GCCACA-5', 884, 3'-GCCAGA-5', 935, 3'-GCCACA-5', 984, 3'-CGCACA-5', 1052, 3'-CGCACA-5', 1136, 3'-TGCACA-5', 1220, 3'-CCCAGT-5', 1250, 3'-CGCAGA-5', 1316, 3'-TGCACT-5', 1372, 3'-CGCAGA-5', 1416, 3'-TGCACT-5', 1472, 3'-CCCACT-5', 1502, 3'-CGCACA-5', 1556, 3'-GGCATT-5', 1702, 3'-CCCAGA-5', 1742, 3'-TGCACA-5', 1822, 3'-TCCACT-5', 1912, 3'-TGCAGA-5', 1937, 3'-GGCACT-5', 1996, 3'-CCCAGT-5', 2024, 3'-TCCACA-5', 2029, 3'-CTCAGT-5', 2060, 3'-TGCAGT-5', 2065, 3'-GCCACT-5', 2072, 3'-TTCAGT-5', 2098, 3'-CTCATA-5', 2176, 3'-TGCATT-5', 2206, 3'-GTCAGA-5', 2222, 3'-CTCAGA-5', 2239, 3'-TTCACT-5', 2304, 3'-TGCAGT-5', 2328, 3'-GTCACT-5', 2425, 3'-GTCAGA-5', 2609, 3'-CTCAGA-5', 2699, 3'-TGCAGA-5', 2721, 3'-CTCAGA-5', 2729, 3'-TGCAGA-5', 2859, 3'-CTCAGA-5', 2866, 3'-CTCATT-5', 2902, 3'-GTCACT-5', 2929, 3'-TTCAGT-5', 2936, 3'-TGCACA-5', 2962, 3'-TGCATT-5', 3072, 3'-CCCAGT-5', 3082, 3'-CCCAGA-5', 3091, 3'-TCCACA-5', 3192, 3'-CTCACA-5', 3209, 3'-GCCAGA-5', 3221, 3'-TGCAGT-5', 3232, 3'-TGCAGT-5', 3281, 3'-CTCACT-5', 3317, 3'-TGCACT-5', 3343, 3'-CCCAGT-5', 3379, 3'-CCCACT-5', 3388, 3'-GGCACA-5', 3409, 3'-TGCAGT-5', 3461, 3'-GGCAGA-5', 3473, 3'-CTCACA-5', 3505, 3'-GCCACA-5', 3705, 3'-TCCAGA-5', 3806, 3'-GTCACA-5', 3822, 3'-TGCAGA-5', 3831, 3'-TCCAGA-5', 3891, 3'-CGCAGA-5', 3916, 3'-GTCACA-5', 3954, 3'-TGCAGT-5', 3962, 3'-GGCACT-5', 4006, 3'-TCCACT-5', 4013, 3'-CTCAGA-5', 4195, 3'-GTCAGT-5', 4271, 3'-CTCATT-5', 4309, 3'-TGCAGA-5', 4317, 3'-CCCAGA-5', 4330, 3'-CTCACT-5', 4338,
- positive strand in the negative direction is SuccessablesInr2+-.bas, looking for 3'-C/G/T-C/G/T-C-A-C/G/T-A/T-5', 59, 3'-GCCATA-5', 39, 3'-TGCATT-5', 152, 3'-GTCACT-5', 208, 3'-GGCACA-5', 266, 3'-GGCACA-5', 518, 3'-GGCACA-5', 960, 3'-GGCAGA-5', 1023, 3'-TGCAGT-5', 1032, 3'-TTCACT-5', 1056, 3'-GGCACA-5', 1116, 3'-CTCACA-5', 1126, 3'-GGCAGA-5', 1314, 3'-TGCAGT-5', 1323, 3'-TGCACT-5', 1347, 3'-TCCAGT-5', 1352, 3'-TCCATT-5', 1378, 3'-CCCAGA-5', 1411, 3'-TGCAGT-5', 1472, 3'-CTCACT-5', 1491, 3'-CCCAGA-5', 1518, 3'-TCCAGT-5', 1532, 3'-TGCACA-5', 1719, 3'-GGCAGA-5', 1967, 3'-TGCAGT-5', 1976, 3'-GCCACT-5', 1995, 3'-TGCACT-5', 2000, 3'-TGCAGT-5', 2083, 3'-GCCAGT-5', 2211, 3'-TGCAGT-5', 2402, 3'-TGCACT-5', 2426, 3'-TCCACT-5', 2632, 3'-GCCAGT-5', 2654, 3'-GGCACA-5', 2665, 3'-TGCAGT-5', 2737, 3'-GCCACT-5', 2756, 3'-GCCATT-5', 3284, 3'-TGCACT-5', 3289, 3'-TGCAGA-5', 3431, 3'-GGCATA-5', 3445, 3'-GGCATA-5', 3451, 3'-GGCAGT-5', 3478, 3'-GGCAGA-5', 3589, 3'-GGCAGT-5', 3600, 3'-GTCAGA-5', 3625, 3'-GGCACA-5', 3632, 3'-CTCAGA-5', 3644, 3'-GCCATT-5', 3686, 3'-TCCACA-5', 3692, 3'-CCCATA-5', 3856, 3'-CTCACA-5', 3965, 3'-GCCAGA-5', 4233, 3'-TGCAGT-5', 4317, 3'-TGCACT-5', 4340, 3'-GCCAGT-5', 4415, 3'-TCCACT-5', 4423, 3'-CCCAGA-5', 4448, 3'-TCCACT-5', 4459, 3'-CCCACT-5', 4485, 3'-TTCACA-5', 4531,
- positive strand in the positive direction is SuccessablesInr2++.bas, looking for 3'-C/G/T-C/G/T-C-A-C/G/T-A/T-5', 40, 3'-TCCAGT-5', 153, 3'-CGCACA-5', 1020, 3'-CCCAGA-5', 1711, 3'-CGCACT-5', 1720, 3'-CCCACA-5', 1803, 3'-CCCAGA-5', 1958, 3'-TCCACA-5', 1969, 3'-GTCAGT-5', 2100, 3'-TCCACT-5', 2128, 3'-TCCAGT-5', 2220, 3'-TCCAGA-5', 2258, 3'-TCCACT-5', 2375, 3'-CGCAGT-5', 2423, 3'-GTCACA-5', 2464, 3'-CCCAGA-5', 2489, 3'-TTCACT-5', 2511, 3'-CGCACT-5', 2555, 3'-GTCAGT-5', 2607, 3'-CTCAGT-5', 2613, 3'-TTCAGT-5', 2618, 3'-TCCATA-5', 2642, 3'-TCCAGA-5', 3019, 3'-CTCAGA-5', 3187, 3'-TGCAGA-5', 3256, 3'-CTCACA-5', 3592, 3'-GCCAGA-5', 3608, 3'-CTCACT-5', 3712, 3'-TCCATT-5', 3731, 3'-TCCAGA-5', 3771, 3'-CCCAGT-5', 3820, 3'-GTCACT-5', 3843, 3'-CTCACT-5', 3876, 3'-TTCAGA-5', 3922, 3'-TCCACT-5', 3934, 3'-GTCACA-5', 3964, 3'-CGCAGA-5', 4056, 3'-TCCAGT-5', 4269, 3'-CTCACT-5', 4350, 3'-CCCACT-5', 4399, 3'-CCCAGA-5', 4414,
- complement, negative strand, negative direction is SuccessablesInr2c--.bas, looking for 3'-A/C/G-A/C/G-G-T-A/C/G-A/T-5', 59, 3'-CGGTAT-5', 39, 3'-ACGTAA-5', 152, 3'-CAGTGA-5', 208, 3'-CCGTGT-5', 266, 3'-CCGTGT-5', 518, 3'-CCGTGT-5', 960, 3'-CCGTCT-5', 1023, 3'-ACGTCA-5', 1032, 3'-AAGTGA-5', 1056, 3'-CCGTGT-5', 1116, 3'-GAGTGT-5', 1126, 3'-CCGTCT-5', 1314, 3'-ACGTCA-5', 1323, 3'-ACGTGA-5', 1347, 3'-AGGTCA-5', 1352, 3'-AGGTAA-5', 1378, 3'-GGGTCT-5', 1411, 3'-ACGTCA-5', 1472, 3'-GAGTGA-5', 1491, 3'-GGGTCT-5', 1518, 3'-AGGTCA-5', 1532, 3'-ACGTGT-5', 1719, 3'-CCGTCT-5', 1967, 3'-ACGTCA-5', 1976, 3'-CGGTGA-5', 1995, 3'-ACGTGA-5', 2000, 3'-ACGTCA-5', 2083, 3'-CGGTCA-5', 2211, 3'-ACGTCA-5', 2402, 3'-ACGTGA-5', 2426, 3'-AGGTGA-5', 2632, 3'-CGGTCA-5', 2654, 3'-CCGTGT-5', 2665, 3'-ACGTCA-5', 2737, 3'-CGGTGA-5', 2756, 3'-CGGTAA-5', 3284, 3'-ACGTGA-5', 3289, 3'-ACGTCT-5', 3431, 3'-CCGTAT-5', 3445, 3'-CCGTAT-5', 3451, 3'-CCGTCA-5', 3478, 3'-CCGTCT-5', 3589, 3'-CCGTCA-5', 3600, 3'-CAGTCT-5', 3625, 3'-CCGTGT-5', 3632, 3'-GAGTCT-5', 3644, 3'-CGGTAA-5', 3686, 3'-AGGTGT-5', 3692, 3'-GGGTAT-5', 3856, 3'-GAGTGT-5', 3965, 3'-CGGTCT-5', 4233, 3'-ACGTCA-5', 4317, 3'-ACGTGA-5', 4340, 3'-CGGTCA-5', 4415, 3'-AGGTGA-5', 4423, 3'-GGGTCT-5', 4448, 3'-AGGTGA-5', 4459, 3'-GGGTGA-5', 4485, 3'-AAGTGT-5', 4531,
- complement, negative strand, positive direction is SuccessablesInr2c-+.bas, looking for 3'-A/C/G-A/C/G-G-T-A/C/G-A/T-5', 40, 3'-AGGTCA-5', 153, 3'-GCGTGT-5', 1020, 3'-GGGTCT-5', 1711, 3'-GCGTGA-5', 1720, 3'-GGGTGT-5', 1803, 3'-GGGTCT-5', 1958, 3'-AGGTGT-5', 1969, 3'-CAGTCA-5', 2100, 3'-AGGTGA-5', 2128, 3'-AGGTCA-5', 2220, 3'-AGGTCT-5', 2258, 3'-AGGTGA-5', 2375, 3'-GCGTCA-5', 2423, 3'-CAGTGT-5', 2464, 3'-GGGTCT-5', 2489, 3'-AAGTGA-5', 2511, 3'-GCGTGA-5', 2555, 3'-CAGTCA-5', 2607, 3'-GAGTCA-5', 2613, 3'-AAGTCA-5', 2618, 3'-AGGTAT-5', 2642, 3'-AGGTCT-5', 3019, 3'-GAGTCT-5', 3187, 3'-ACGTCT-5', 3256, 3'-GAGTGT-5', 3592, 3'-CGGTCT-5', 3608, 3'-GAGTGA-5', 3712, 3'-AGGTAA-5', 3731, 3'-AGGTCT-5', 3771, 3'-GGGTCA-5', 3820, 3'-CAGTGA-5', 3843, 3'-GAGTGA-5', 3876, 3'-AAGTCT-5', 3922, 3'-AGGTGA-5', 3934, 3'-CAGTGT-5', 3964, 3'-GCGTCT-5', 4056, 3'-AGGTCA-5', 4269, 3'-GAGTGA-5', 4350, 3'-GGGTGA-5', 4399, 3'-GGGTCT-5', 4414,
- complement, positive strand, negative direction is SuccessablesInr2c+-.bas, looking for 3'-A/C/G-A/C/G-G-T-A/C/G-A/T-5', 44, 3'-AGGTAT-5', 179, 3'-GGGTCA-5', 206, 3'-GAGTCT-5', 278, 3'-CAGTGA-5', 299, 3'-AAGTGT-5', 322, 3'-AGGTCA-5', 439, 3'-ACGTAA-5', 533, 3'-AGGTCA-5', 568, 3'-AGGTCA-5', 576, 3'-AGGTCA-5', 712, 3'-CCGTCT-5', 754, 3'-CGGTGA-5', 868, 3'-CAGTGA-5', 1034, 3'-GGGTGA-5', 1049, 3'-GAGTGA-5', 1077, 3'-CCGTGT-5', 1220, 3'-CAGTGA-5', 1325, 3'-CAGTCT-5', 1354, 3'-GAGTCT-5', 1444, 3'-CCGTCA-5', 1511, 3'-ACGTCT-5', 1774, 3'-CAGTGA-5', 1978, 3'-CAGTGT-5', 2085, 3'-AGGTCA-5', 2248, 3'-CAGTGA-5', 2404, 3'-GAGTGA-5', 2447, 3'-AGGTCA-5', 2585, 3'-CAGTGT-5', 2603, 3'-CAGTGT-5', 2656, 3'-CAGTGA-5', 2739, 3'-AAGTGT-5', 2860, 3'-AGGTGA-5', 3144, 3'-GGGTGT-5', 3184, 3'-AAGTGA-5', 3410, 3'-CAGTAA-5', 3480, 3'-AGGTGA-5', 3825, 3'-GAGTAT-5', 3829, 3'-GAGTAA-5', 3891, 3'-AAGTGT-5', 3939, 3'-CAGTGA-5', 4200, 3'-AGGTCA-5', 4307, 3'-CAGTGA-5', 4319, 3'-GGGTGA-5', 4353, 3'-CAGTGT-5', 4359,
- complement, positive strand, positive direction is SuccessablesInr2c++.bas, looking for 3'-A/C/G-A/C/G-G-T-A/C/G-A/T-5', 87, 3'-AGGTCT-5', 15, 3'-CCGTAA-5', 22, 3'-CAGTGT-5', 155, 3'-GGGTCT-5', 204, 3'-CGGTGT-5', 343, 3'-GCGTCT-5', 396, 3'-ACGTCT-5', 438, 3'-GGGTCT-5', 468, 3'-ACGTGT-5', 548, 3'-AGGTGT-5', 632, 3'-GCGTGA-5', 686, 3'-GCGTGT-5', 800, 3'-CGGTCT-5', 835, 3'-CGGTGT-5', 884, 3'-CGGTCT-5', 935, 3'-CGGTGT-5', 984, 3'-GCGTGT-5', 1052, 3'-GCGTGT-5', 1136, 3'-ACGTGT-5', 1220, 3'-GGGTCA-5', 1250, 3'-GCGTCT-5', 1316, 3'-ACGTGA-5', 1372, 3'-GCGTCT-5', 1416, 3'-ACGTGA-5', 1472, 3'-GGGTGA-5', 1502, 3'-GCGTGT-5', 1556, 3'-CCGTAA-5', 1702, 3'-GGGTCT-5', 1742, 3'-ACGTGT-5', 1822, 3'-AGGTGA-5', 1912, 3'-ACGTCT-5', 1937, 3'-CCGTGA-5', 1996, 3'-GGGTCA-5', 2024, 3'-AGGTGT-5', 2029, 3'-GAGTCA-5', 2060, 3'-ACGTCA-5', 2065, 3'-CGGTGA-5', 2072, 3'-AAGTCA-5', 2098, 3'-GAGTAT-5', 2176, 3'-ACGTAA-5', 2206, 3'-CAGTCT-5', 2222, 3'-GAGTCT-5', 2239, 3'-AAGTGA-5', 2304, 3'-ACGTCA-5', 2328, 3'-CAGTGA-5', 2425, 3'-CAGTCT-5', 2609, 3'-GAGTCT-5', 2699, 3'-ACGTCT-5', 2721, 3'-GAGTCT-5', 2729, 3'-ACGTCT-5', 2859, 3'-GAGTCT-5', 2866, 3'-GAGTAA-5', 2902, 3'-CAGTGA-5', 2929, 3'-AAGTCA-5', 2936, 3'-ACGTGT-5', 2962, 3'-ACGTAA-5', 3072, 3'-GGGTCA-5', 3082, 3'-GGGTCT-5', 3091, 3'-AGGTGT-5', 3192, 3'-GAGTGT-5', 3209, 3'-CGGTCT-5', 3221, 3'-ACGTCA-5', 3232, 3'-ACGTCA-5', 3281, 3'-GAGTGA-5', 3317, 3'-ACGTGA-5', 3343, 3'-GGGTCA-5', 3379, 3'-GGGTGA-5', 3388, 3'-CCGTGT-5', 3409, 3'-ACGTCA-5', 3461, 3'-CCGTCT-5', 3473, 3'-GAGTGT-5', 3505, 3'-CGGTGT-5', 3705, 3'-AGGTCT-5', 3806, 3'-CAGTGT-5', 3822, 3'-ACGTCT-5', 3831, 3'-AGGTCT-5', 3891, 3'-GCGTCT-5', 3916, 3'-CAGTGT-5', 3954, 3'-ACGTCA-5', 3962, 3'-CCGTGA-5', 4006, 3'-AGGTGA-5', 4013, 3'-GAGTCT-5', 4195, 3'-CAGTCA-5', 4271, 3'-GAGTAA-5', 4309, 3'-ACGTCT-5', 4317, 3'-GGGTCT-5', 4330, 3'-GAGTGA-5', 4338,
- inverse complement, negative strand, negative direction is SuccessablesInr2ci--.bas, looking for 3'-A/T-A/C/G-T-G-A/C/G-A/C/G-5', 46, 3'-TCTGAC-5', 16, 3'-TGTGGA-5', 62, 3'-TGTGCA-5', 342, 3'-TGTGCA-5', 531, 3'-AGTGCG-5', 663, 3'-TGTGGG-5', 749, 3'-TCTGAG-5', 916, 3'-TGTGCG-5', 963, 3'-ACTGAA-5', 1052, 3'-AGTGAG-5', 1057, 3'-TCTGAG-5', 1082, 3'-TGTGGA-5', 1129, 3'-AGTGGA-5', 1171, 3'-AATGAA-5', 1298, 3'-TCTGAG-5', 1403, 3'-AGTGAC-5', 1492, 3'-TGTGAA-5', 1544, 3'-TCTGAA-5', 1617, 3'-AGTGCA-5', 1772, 3'-TCTGAC-5', 1934, 3'-AGTGCG-5', 1991, 3'-TCTGAG-5', 2026, 3'-TATGAC-5', 2162, 3'-ACTGGC-5', 2190, 3'-AGTGCG-5', 2207, 3'-TGTGAA-5', 2551, 3'-AGTGAA-5', 2578, 3'-ACTGAG-5', 2787, 3'-TATGGA-5', 2994, 3'-AGTGGG-5', 3057, 3'-AGTGAA-5', 3101, 3'-AGTGAA-5', 3240, 3'-AGTGCG-5', 3280, 3'-TCTGAC-5', 3425, 3'-TATGAC-5', 3541, 3'-TATGCG-5', 3547, 3'-TATGGA-5', 3859, 3'-TGTGGA-5', 3968, 3'-TGTGAA-5', 3983, 3'-AGTGAA-5', 4010, 3'-TCTGAG-5', 4054, 3'-AGTGAA-5', 4161, 3'-TCTGGG-5', 4205, 3'-TCTGCA-5', 4236, 3'-TGTGAC-5', 4336, 3'-TCTGGG-5', 4366,
- inverse complement, negative strand, positive direction is SuccessablesInr2ci-+.bas, looking for 3'-A/T-A/C/G-T-G-A/C/G-A/C/G-5', 94, 3'-AGTGGG-5', 54, 3'-TCTGCA-5', 224, 3'-TGTGAA-5', 231, 3'-ACTGCC-5', 238, 3'-TCTGAG-5', 256, 3'-TCTGGA-5', 271, 3'-ACTGGG-5', 348, 3'-AGTGCG-5', 497, 3'-AGTGCG-5', 581, 3'-AGTGCG-5', 665, 3'-ACTGCG-5', 749, 3'-TGTGGC-5', 819, 3'-ACTGCC-5', 901, 3'-TGTGGC-5', 919, 3'-ACTGCG-5', 1001, 3'-TGTGGC-5', 1023, 3'-AGTGCG-5', 1085, 3'-AGTGCG-5', 1160, 3'-AGTGCG-5', 1169, 3'-AGTGCG-5', 1253, 3'-ACTGAG-5', 1287, 3'-AATGCG-5', 1321, 3'-TCTGGC-5', 1377, 3'-TCTGCG-5', 1396, 3'-AATGCG-5', 1421, 3'-TCTGGC-5', 1477, 3'-TCTGCG-5', 1496, 3'-ACTGCA-5', 1505, 3'-AGTGCG-5', 1589, 3'-AGTGCG-5', 1725, 3'-AGTGCA-5', 1786, 3'-TGTGGA-5', 1806, 3'-TCTGGG-5', 1865, 3'-ACTGGG-5', 1954, 3'-TGTGGC-5', 1972, 3'-TCTGGC-5', 1993, 3'-AGTGCA-5', 2063, 3'-AGTGGC-5', 2068, 3'-TATGGC-5', 2160, 3'-ACTGCA-5', 2204, 3'-AGTGCA-5', 2326, 3'-TGTGCA-5', 2681, 3'-AGTGGA-5', 2712, 3'-ACTGCC-5', 2823, 3'-AATGAC-5', 2842, 3'-TCTGCA-5', 2857, 3'-TCTGGC-5', 2884, 3'-AATGGG-5', 2911, 3'-TCTGAC-5', 2944, 3'-TCTGAG-5', 2951, 3'-TGTGCA-5', 2960, 3'-TCTGGC-5', 2984, 3'-TCTGAG-5', 3007, 3'-AGTGCC-5', 3011, 3'-TATGAC-5', 3028, 3'-TCTGCA-5', 3061, 3'-AATGCA-5', 3070, 3'-ACTGGC-5', 3118, 3'-TCTGAG-5', 3124, 3'-TATGGA-5', 3163, 3'-AATGGG-5', 3169, 3'-AGTGCC-5', 3235, 3'-TATGAG-5', 3261, 3'-TCTGCA-5', 3268, 3'-TCTGCA-5', 3279, 3'-ACTGCA-5', 3320, 3'-ACTGGC-5', 3346, 3'-TCTGCC-5', 3359, 3'-TCTGGC-5', 3406, 3'-AATGCC-5', 3431, 3'-TGTGGA-5', 3437, 3'-AATGAA-5', 3442, 3'-AATGAG-5', 3446, 3'-AGTGGG-5', 3450, 3'-AGTGCA-5', 3464, 3'-AATGAC-5', 3568, 3'-TGTGAA-5', 3595, 3'-AGTGAC-5', 3713, 3'-ACTGAG-5', 3736, 3'-AATGAC-5', 3783, 3'-AATGAA-5', 3836, 3'-AGTGAG-5', 3877, 3'-TGTGAG-5', 3904, 3'-TCTGAA-5', 3925, 3'-TGTGCA-5', 3960, 3'-TGTGAC-5', 3972, 3'-AGTGGG-5', 4041, 3'-ACTGAA-5', 4090, 3'-AATGAG-5', 4095, 3'-AGTGCC-5', 4274, 3'-ACTGCA-5', 4341, 3'-AGTGAG-5', 4351, 3'-TGTGGG-5', 4395, 3'-TCTGGG-5', 4417,
- inverse complement, positive strand, negative direction is SuccessablesInr2ci+-.bas, looking for 3'-A/T-A/C/G-T-G-A/C/G-A/C/G-5', 54, 3'-ACTGAA-5', 18, 3'-TATGGG-5', 78, 3'-ACTGAA-5', 131, 3'-TATGAG-5', 275, 3'-AGTGAG-5', 300, 3'-ACTGAC-5', 308, 3'-AGTGCG-5', 447, 3'-AGTGAA-5', 472, 3'-AGTGGA-5', 523, 3'-AGTGAG-5', 1035, 3'-AGTGAG-5', 1078, 3'-AGTGGC-5', 1121, 3'-AGTGAG-5', 1326, 3'-TCTGGG-5', 1357, 3'-AGTGCA-5', 1470, 3'-ACTGCA-5', 1494, 3'-AGTGCA-5', 1535, 3'-AATGAA-5', 1581, 3'-AATGCC-5', 1634, 3'-TATGGC-5', 1743, 3'-ACTGAG-5', 1936, 3'-AATGGC-5', 1949, 3'-AGTGAG-5', 1979, 3'-ACTGCA-5', 1998, 3'-TGTGGC-5', 2066, 3'-AATGAC-5', 2188, 3'-AGTGAG-5', 2405, 3'-ACTGCA-5', 2424, 3'-AGTGAG-5', 2448, 3'-TGTGGC-5', 2606, 3'-AGTGAG-5', 2740, 3'-ACTGCA-5', 2759, 3'-TGTGCA-5', 2863, 3'-AATGGC-5', 3005, 3'-TGTGAG-5', 3268, 3'-AGTGAC-5', 3411, 3'-TGTGCA-5', 3429, 3'-TGTGCC-5', 3561, 3'-AATGGG-5', 3660, 3'-TGTGGG-5', 3712, 3'-ACTGGG-5', 3750, 3'-AATGCA-5', 3771, 3'-TCTGGA-5', 3836, 3'-ACTGCC-5', 3852, 3'-TGTGGC-5', 3960, 3'-AGTGAG-5', 4050, 3'-TGTGAG-5', 4093, 3'-AGTGAG-5', 4201, 3'-ACTGCA-5', 4315, 3'-AGTGAG-5', 4320, 3'-ACTGCA-5', 4330, 3'-ACTGCA-5', 4338, 3'-TGTGAG-5', 4362, 3'-AATGAG-5', 4556,
- inverse complement, positive strand, positive direction is SuccessablesInr2ci++.bas, looking for 3'-A/T-A/C/G-T-G-A/C/G-A/C/G-5', 47, 3'-TCTGAC-5', 236, 3'-TGTGAC-5', 346, 3'-TCTGCC-5', 399, 3'-TCTGGC-5', 441, 3'-AATGAA-5', 525, 3'-TGTGCA-5', 569, 3'-TGTGCG-5', 803, 3'-TGTGCG-5', 887, 3'-TGTGCG-5', 987, 3'-TGTGAC-5', 1139, 3'-TGTGCC-5', 1223, 3'-TGTGCC-5', 1559, 3'-ACTGGG-5', 1663, 3'-TGTGCC-5', 1698, 3'-TCTGAA-5', 1745, 3'-AATGGG-5', 1889, 3'-ACTGGC-5', 2214, 3'-AGTGGA-5', 2248, 3'-AGTGAG-5', 2305, 3'-AGTGGG-5', 2313, 3'-AGTGAC-5', 2341, 3'-TCTGAA-5', 2417, 3'-TGTGGA-5', 2431, 3'-TATGAA-5', 2740, 3'-TCTGGA-5', 2862, 3'-AGTGAC-5', 2930, 3'-ACTGAA-5', 2946, 3'-TGTGGG-5', 2965, 3'-ACTGAA-5', 3030, 3'-AGTGCA-5', 3254, 3'-AGTGAC-5', 3318, 3'-TGTGAG-5', 3508, 3'-TGTGGG-5', 3533, 3'-TCTGGA-5', 3551, 3'-AGTGGG-5', 3613, 3'-AGTGCC-5', 3748, 3'-ACTGGA-5', 3785, 3'-ACTGGA-5', 4019, 3'-AGTGAC-5', 4088, 3'-AGTGAG-5', 4127, 3'-AGTGGG-5', 4204, 3'-ACTGGG-5', 4217, 3'-TGTGCC-5', 4259, 3'-TCTGCG-5', 4320, 3'-AGTGGG-5', 4326, 3'-TGTGAG-5', 4335, 3'-AGTGAC-5', 4339,
- inverse, negative strand, negative direction, is SuccessablesInr2i--.bas, looking for 3'-A/T-C/G/T-A-C-C/G/T-C/G/T-5', 54, 3'-TGACTT-5', 18, 3'-ATACCC-5', 78, 3'-TGACTT-5', 131, 3'-ATACTC-5', 275, 3'-TCACTC-5', 300, 3'-TGACTG-5', 308, 3'-TCACGC-5', 447, 3'-TCACTT-5', 472, 3'-TCACCT-5', 523, 3'-TCACTC-5', 1035, 3'-TCACTC-5', 1078, 3'-TCACCG-5', 1121, 3'-TCACTC-5', 1326, 3'-AGACCC-5', 1357, 3'-TCACGT-5', 1470, 3'-TGACGT-5', 1494, 3'-TCACGT-5', 1535, 3'-TTACTT-5', 1581, 3'-TTACGG-5', 1634, 3'-ATACCG-5', 1743, 3'-TGACTC-5', 1936, 3'-TTACCG-5', 1949, 3'-TCACTC-5', 1979, 3'-TGACGT-5', 1998, 3'-ACACCG-5', 2066, 3'-TTACTG-5', 2188, 3'-TCACTC-5', 2405, 3'-TGACGT-5', 2424, 3'-TCACTC-5', 2448, 3'-ACACCG-5', 2606, 3'-TCACTC-5', 2740, 3'-TGACGT-5', 2759, 3'-ACACGT-5', 2863, 3'-TTACCG-5', 3005, 3'-ACACTC-5', 3268, 3'-TCACTG-5', 3411, 3'-ACACGT-5', 3429, 3'-ACACGG-5', 3561, 3'-TTACCC-5', 3660, 3'-ACACCC-5', 3712, 3'-TGACCC-5', 3750, 3'-TTACGT-5', 3771, 3'-AGACCT-5', 3836, 3'-TGACGG-5', 3852, 3'-ACACCG-5', 3960, 3'-TCACTC-5', 4050, 3'-ACACTC-5', 4093, 3'-TCACTC-5', 4201, 3'-TGACGT-5', 4315, 3'-TCACTC-5', 4320, 3'-TGACGT-5', 4330, 3'-TGACGT-5', 4338, 3'-ACACTC-5', 4362, 3'-TTACTC-5', 4556,
- inverse, negative strand, positive direction, is SuccessablesInr2i-+.bas, looking for 3'-A/T-C/G/T-A-C-C/G/T-C/G/T-5', 47 , 3'-AGACTG-5', 236 , 3'-ACACTG-5', 346 , 3'-AGACGG-5', 399 , 3'-AGACCG-5', 441 , 3'-TTACTT-5', 525 , 3'-ACACGT-5', 569 , 3'-ACACGC-5', 803 , 3'-ACACGC-5', 887 , 3'-ACACGC-5', 987 , 3'-ACACTG-5', 1139 , 3'-ACACGG-5', 1223 , 3'-ACACGG-5', 1559 , 3'-TGACCC-5', 1663 , 3'-ACACGG-5', 1698 , 3'-AGACTT-5', 1745 , 3'-TTACCC-5', 1889 , 3'-TGACCG-5', 2214 , 3'-TCACCT-5', 2248 , 3'-TCACTC-5', 2305 , 3'-TCACCC-5', 2313 , 3'-TCACTG-5', 2341 , 3'-AGACTT-5', 2417 , 3'-ACACCT-5', 2431 , 3'-ATACTT-5', 2740 , 3'-AGACCT-5', 2862 , 3'-TCACTG-5', 2930 , 3'-TGACTT-5', 2946 , 3'-ACACCC-5', 2965 , 3'-TGACTT-5', 3030 , 3'-TCACGT-5', 3254 , 3'-TCACTG-5', 3318 , 3'-ACACTC-5', 3508 , 3'-ACACCC-5', 3533 , 3'-AGACCT-5', 3551 , 3'-TCACCC-5', 3613 , 3'-TCACGG-5', 3748 , 3'-TGACCT-5', 3785 , 3'-TGACCT-5', 4019 , 3'-TCACTG-5', 4088 , 3'-TCACTC-5', 4127 , 3'-TCACCC-5', 4204 , 3'-TGACCC-5', 4217 , 3'-ACACGG-5', 4259 , 3'-AGACGC-5', 4320 , 3'-TCACCC-5', 4326 , 3'-ACACTC-5', 4335 , 3'-TCACTG-5', 4339,
- inverse, positive strand, negative direction, is SuccessablesInr2i+-.bas, looking for 3'-A/T-C/G/T-A-C-C/G/T-C/G/T-5', 46, 3'-AGACTG-5', 16, 3'-ACACCT-5', 62, 3'-ACACGT-5', 342, 3'-ACACGT-5', 531, 3'-TCACGC-5', 663, 3'-ACACCC-5', 749, 3'-AGACTC-5', 916, 3'-ACACGC-5', 963, 3'-TGACTT-5', 1052, 3'-TCACTC-5', 1057, 3'-AGACTC-5', 1082, 3'-ACACCT-5', 1129, 3'-TCACCT-5', 1171, 3'-TTACTT-5', 1298, 3'-AGACTC-5', 1403, 3'-TCACTG-5', 1492, 3'-ACACTT-5', 1544, 3'-AGACTT-5', 1617, 3'-TCACGT-5', 1772, 3'-AGACTG-5', 1934, 3'-TCACGC-5', 1991, 3'-AGACTC-5', 2026, 3'-ATACTG-5', 2162, 3'-TGACCG-5', 2190, 3'-TCACGC-5', 2207, 3'-ACACTT-5', 2551, 3'-TCACTT-5', 2578, 3'-TGACTC-5', 2787, 3'-ATACCT-5', 2994, 3'-TCACCC-5', 3057, 3'-TCACTT-5', 3101, 3'-TCACTT-5', 3240, 3'-TCACGC-5', 3280, 3'-AGACTG-5', 3425, 3'-ATACTG-5', 3541, 3'-ATACGC-5', 3547, 3'-ATACCT-5', 3859, 3'-ACACCT-5', 3968, 3'-ACACTT-5', 3983, 3'-TCACTT-5', 4010, 3'-AGACTC-5', 4054, 3'-TCACTT-5', 4161, 3'-AGACCC-5', 4205, 3'-AGACGT-5', 4236, 3'-ACACTG-5', 4336, 3'-AGACCC-5', 4366,
- inverse, positive strand, positive direction, is SuccessablesInr2i++.bas, looking for 3'-A/T-C/G/T-A-C-C/G/T-C/G/T-5', 94, 3'-TCACCC-5', 54, 3'-AGACGT-5', 224, 3'-ACACTT-5', 231, 3'-TGACGG-5', 238, 3'-AGACTC-5', 256, 3'-AGACCT-5', 271, 3'-TGACCC-5', 348, 3'-TCACGC-5', 497, 3'-TCACGC-5', 581, 3'-TCACGC-5', 665, 3'-TGACGC-5', 749, 3'-ACACCG-5', 819, 3'-TGACGG-5', 901, 3'-ACACCG-5', 919, 3'-TGACGC-5', 1001, 3'-ACACCG-5', 1023, 3'-TCACGC-5', 1085, 3'-TCACGC-5', 1160, 3'-TCACGC-5', 1169, 3'-TCACGC-5', 1253, 3'-TGACTC-5', 1287, 3'-TTACGC-5', 1321, 3'-AGACCG-5', 1377, 3'-AGACGC-5', 1396, 3'-TTACGC-5', 1421, 3'-AGACCG-5', 1477, 3'-AGACGC-5', 1496, 3'-TGACGT-5', 1505, 3'-TCACGC-5', 1589, 3'-TCACGC-5', 1725, 3'-TCACGT-5', 1786, 3'-ACACCT-5', 1806, 3'-AGACCC-5', 1865, 3'-TGACCC-5', 1954, 3'-ACACCG-5', 1972, 3'-AGACCG-5', 1993, 3'-TCACGT-5', 2063, 3'-TCACCG-5', 2068, 3'-ATACCG-5', 2160, 3'-TGACGT-5', 2204, 3'-TCACGT-5', 2326, 3'-ACACGT-5', 2681, 3'-TCACCT-5', 2712, 3'-TGACGG-5', 2823, 3'-TTACTG-5', 2842, 3'-AGACGT-5', 2857, 3'-AGACCG-5', 2884, 3'-TTACCC-5', 2911, 3'-AGACTG-5', 2944, 3'-AGACTC-5', 2951, 3'-ACACGT-5', 2960, 3'-AGACCG-5', 2984, 3'-AGACTC-5', 3007, 3'-TCACGG-5', 3011, 3'-ATACTG-5', 3028, 3'-AGACGT-5', 3061, 3'-TTACGT-5', 3070, 3'-TGACCG-5', 3118, 3'-AGACTC-5', 3124, 3'-ATACCT-5', 3163, 3'-TTACCC-5', 3169, 3'-TCACGG-5', 3235, 3'-ATACTC-5', 3261, 3'-AGACGT-5', 3268, 3'-AGACGT-5', 3279, 3'-TGACGT-5', 3320, 3'-TGACCG-5', 3346, 3'-AGACGG-5', 3359, 3'-AGACCG-5', 3406, 3'-TTACGG-5', 3431, 3'-ACACCT-5', 3437, 3'-TTACTT-5', 3442, 3'-TTACTC-5', 3446, 3'-TCACCC-5', 3450, 3'-TCACGT-5', 3464, 3'-TTACTG-5', 3568, 3'-ACACTT-5', 3595, 3'-TCACTG-5', 3713, 3'-TGACTC-5', 3736, 3'-TTACTG-5', 3783, 3'-TTACTT-5', 3836, 3'-TCACTC-5', 3877, 3'-ACACTC-5', 3904, 3'-AGACTT-5', 3925, 3'-ACACGT-5', 3960, 3'-ACACTG-5', 3972, 3'-TCACCC-5', 4041, 3'-TGACTT-5', 4090, 3'-TTACTC-5', 4095, 3'-TCACGG-5', 4274, 3'-TGACGT-5', 4341, 3'-TCACTC-5', 4351, 3'-ACACCC-5', 4395, 3'-AGACCC-5', 4417.
Verifications[edit | edit source]
To verify that your sampling has explored something, you may need a control group. Perhaps where, when, or without your entity, source, or object may serve.
Another verifier is reproducibility. Can you replicate something about your entity in your laboratory more than 3 times. Five times is usually a beginning number to provide statistics (data) about it.
For an apparent one time or perception event, document or record as much information coincident as possible. Was there a butterfly nearby?
Has anyone else perceived the entity and recorded something about it?
Gene ID: 1, includes the nucleotides between neighboring genes and A1BG. These nucleotides can be loaded into files from either gene toward A1BG, and from template and coding strands. These nucleotide sequences can be found in Gene transcriptions/A1BG. Copying the above discovered CRE boxes and putting the sequences in "⌘F" locates these sequences in the same nucleotide positions as found by the computer programs.
Core promoter initiator elements[edit | edit source]
From the first nucleotide just after ZSCAN22 to the first nucleotide just before A1BG are 4460 nucleotides. The core promoter on this side of A1BG extends from approximately 4425 to the possible transcription start site at nucleotide number 4460.
For the consensus sequence YYANWYY: there is the following Inr in the core promoter, negative strand, negative direction: 3'-TTACTCC-5' at 4557.
For the consensus sequence BBCABW: there are no Inr in the core promoter, negative strand, negative direction.
For the consensus sequence YYANWYY: there are four Inrs in the core promoter, positive strand, negative direction: 3'-CCACTCC-5' at 4425, 3'-CCACTTT-5' at 4461, 3'-TCACATT-5' at 4533, and 3'-TTAATTC-5' at 4542.
For the consensus sequence BBCABW: there are five Inrs in the core promoter, positive strand, negative direction: 3'-TCCACT-5', 4423, 3'-CCCAGA-5', 4448, 3'-TCCACT-5', 4459, 3'-CCCACT-5', 4485, 3'-TTCACA-5', 4531.
From the first nucleotide just after ZNF497 to the first nucleotide just before A1BG are 858 nucleotides. The core promoter on this side of A1BG extends from approximately 824 to the possible transcription start site at nucleotide number 858. Nucleotides (nts) have been added from ZNF497 to A1BG. The TSS for A1BG is now at 4300 nts from just on the other side of ZNF497. The core promoter should now be from 4266 to 4300.
For the consensus sequence YYANWYY: there is the following Inr in the core promoter, negative strand, positive direction: 3'-CTGCACC-5' at 4343.
For the consensus sequence BBCABW: there are five Inr in the core promoter, negative strand, positive direction: 3'-GTCAGT-5', 4271, 3'-CTCATT-5', 4309, 3'-TGCAGA-5', 4317, 3'-CCCAGA-5', 4330, 3'-CTCACT-5', 4338.
For the consensus sequence YYANWYY: there are two Inrs in the core promoter, positive strand, positive direction: 3'-CCACTCC-5' at 4401 and 3'-CCAGACC-5' at 4416.
For the consensus sequence BBCABW: there are four Inrs in the core promoter, positive strand, positive direction: 3'-TCCAGT-5', 4269, 3'-CTCACT-5', 4350, 3'-CCCACT-5', 4399, 3'-CCCAGA-5', 4414.
Proximal promoter initiator elements[edit | edit source]
The proximal promoter begins about nucleotide number 4210 in the negative direction.
There are eight YYANWYY Inrs on the negative strand in the negative direction: 3'-TCACTCT-5' at 4202, 3'-TCGGTCT-5' at 4233, 3'-CTGCACC-5' at 4238, 3'-TCGGACC-5' at 4300, 3'-CCAGTTT-5' at 4309, 3'-TCGGACC-5' at 4349, 3'-TCACACT-5' at 4361, and 3'-TTACTCC-5' at 4557.
There are five BBCABW Inrs on the negative strand in the negative direction: 3'-GTCACT-5', 4200, 3'-TCCAGT-5', 4307, 3'-GTCACT-5', 4319, 3'-CCCACT-5', 4353, 3'-GTCACA-5', 4359.
There are seven YYANWYY Inrs on the positive strand in the negative direction: 3'-CCGGACT-5' at 4327, 3'-CTGCACT-5' at 4340, 3'-CCAGTTC-5' at 4417, 3'-CCACTCC-5' at 4425, 3'-CCACTTT-5' at 4461, 3'-TCACATT-5' at 4533, and 3'-TTAATTC-5' at 4542.
There are nine BBCABW Inrs on the positive strand in the negative direction: 3'-GCCAGA-5', 4233, 3'-TGCAGT-5', 4317, 3'-TGCACT-5', 4340, 3'-GCCAGT-5', 4415, 3'-TCCACT-5', 4423, 3'-CCCAGA-5', 4448, 3'-TCCACT-5', 4459, 3'-CCCACT-5', 4485, 3'-TTCACA-5', 4531.
The proximal promoter begins about nucleotide number 4195 in the positive direction.
There is one YYANWYY Inr on the negative strand in the positive direction: 3'-CTGCACC-5' at 4343.
There is six BBCABW Inr on the negative strand in the positive direction: 3'-CTCAGA-5', 4195, 3'-GTCAGT-5', 4271, 3'-CTCATT-5', 4309, 3'-TGCAGA-5', 4317, 3'-CCCAGA-5', 4330, 3'-CTCACT-5', 4338.
There is two YYANWYY Inrs on the positive strand in the positive direction: 3'-CCACTCC-5' at 4401 and 3'-CCAGACC-5' at 4416.
There is four BBCABW Inrs on the positive strand in the positive direction: 3'-TCCAGT-5', 4269, 3'-CTCACT-5', 4350, 3'-CCCACT-5', 4399, 3'-CCCAGA-5', 4414.
Distal promoter initiator elements[edit | edit source]
Using an estimate of 2 knts, a distal promoter to A1BG would be expected after nucleotide number 2460 in the negative direction.
These are 45 YYANWYY Inrs on the negative strand in the negative direction: 3'-TCGTTTT-5' at 2476, 3'-TTGTTTT-5' at 2490, 3'-TCATTCT-5' at 2503, 3'-CCGGTCC-5' at 2519, 3'-CCAGTCC-5' at 2587, 3'-TCACACC-5' at 2605, 3'-TTGTACC-5' at 2614, 3'-CCACTTT-5' at 2619, 3'-TCACACC-5' at 2658, 3'-TTGGACC-5' at 2720, 3'-TCGGACC-5' at 2770, 3'-TCGTACT-5' at 2784, 3'-TTGATTC-5' at 2914, 3'-CCGATTT-5' at 3009, 3'-TTGATTC-5' at 3031, 3'-CCGCACC-5' at 3047, 3'-TCGGACC-5' at 3128, 3'-TTGTTCC-5' at 3141, 3'-CCACTTT-5' at 3146, 3'-TTGTATT-5' at 3169, 3'-CCACACC-5' at 3186, 3'-TCGGTTC-5' at 3273, 3'-TCGGACC-5' at 3298, 3'-TTGTTCT-5' at 3307, 3'-TCGTTTT-5' at 3313, 3'-TTGTTCT-5' at 3340, 3'-TCGTTCT-5' at 3374, 3'-CCGAACT-5' at 3401, 3'-CCGTATC-5' at 3446, 3'-TTGATCT-5' at 3463, 3'-TTGGTCT-5' at 3486, 3'-CTGTTCT-5' at 3759, 3'-CTACACC-5' at 3810, 3'-CTGGTCC-5' at 3871, 3'-TCATTCT-5' at 3893, 3'-CTACTTT-5' at 3922, 3'-CCGGTCC-5' at 3951, 3'-TCGGACC-5' at 4037, 3'-TTGTATC-5' at 4046, 3'-TCACTCT-5' at 4051, 3'-TTACACT-5' at 4092, 3'-CCGGTCC-5' at 4102, 3'-CCGTACC-5' at 4107, 3'-CCGGTCC-5' at 4170, 3'-TCGAACC-5' at 4188, and 3'-TCACTCT-5' at 4202.
These are 14 BBCABW Inrs on the negative strand in the negative direction: 3'-CTCACT-5', 2447, 3'-TCCAGT-5', 2585, 3'-GTCACA-5', 2603, 3'-GTCACA-5', 2656, 3'-GTCACT-5', 2739, 3'-TTCACA-5', 2860, 3'-TCCACT-5', 3144, 3'-CCCACA-5', 3184, 3'-TTCACT-5', 3410, 3'-GTCATT-5', 3480, 3'-TCCACT-5', 3825, 3'-CTCATA-5', 3829, 3'-CTCATT-5', 3891, 3'-TTCACA-5', 3939, 3'-GTCACT-5', 4200.
These are 12 YYANWYY Inrs on the positive strand in the negative direction: 3'-TTGAATC-5' at 2708, 3'-TTGAACC-5' at 2717, 3'-CTGCACC-5' at 2761, 3'-TTGAACC-5' at 3245, 3'-TTGCACT-5' at 3289, 3'-CCAGATC-5' at 3488, 3'-CTGCTCC-5' at 3582, 3'-CCATTTC-5' at 3688, 3'-CTGGACT-5' at 3747, 3'-CTGAACC-5' at 3784, 3'-CCATACC-5' at 3858, and 3'-TCACACC-5' at 3967.
These are 21 BBCABW Inrs on the positive strand in the negative direction: 3'-TCCACT-5', 2632, 3'-GCCAGT-5', 2654, 3'-GGCACA-5', 2665, 3'-TGCAGT-5', 2737, 3'-GCCACT-5', 2756, 3'-GCCATT-5', 3284, 3'-TGCACT-5', 3289, 3'-TGCAGA-5', 3431, 3'-GGCATA-5', 3445, 3'-GGCATA-5', 3451, 3'-GGCAGT-5', 3478, 3'-GGCAGA-5', 3589, 3'-GGCAGT-5', 3600, 3'-GTCAGA-5', 3625, 3'-GGCACA-5', 3632, 3'-CTCAGA-5', 3644, 3'-GCCATT-5', 3686, 3'-TCCACA-5', 3692, 3'-CCCATA-5', 3856, 3'-CTCACA-5', 3965, 3'-GCCAGA-5', 4233.
Using an estimate of 2 knts, a distal promoter to A1BG would be expected after nucleotide number 2300 in the positive direction.
These are 28 YYANWYY Inrs on the negative strand in the positive direction: 3'-TCACTCT-5' at 2306, 3'-CTACACC-5' at 2430, 3'-CTAATTT-5' at 2440, 3'-CCGCACC-5' at 2566, 3'-TTATACC-5' at 2590, 3'-CCACACC-5' at 2602, 3'-CCACACT-5' at 2636, 3'-TCAGATT-5' at 2868, 3'-CTGCTCC-5' at 2978, 3'-CCAGTCC-5' at 2998, 3'-CCAGTCC-5' at 3084, 3'-CTGGTCT-5' at 3245, 3'-TCGCTCT-5' at 3276, 3'-CTGGTCT-5' at 3299, 3'-CTGCTCC-5' at 3309, 3'-CTGCACC-5' at 3322, 3'-CCGCATC-5' at 3328, 3'-TTGCACT-5' at 3343, 3'-CTGTTCC-5' at 3352, 3'-TTGCATC-5' at 3402, 3'-TCACACT-5' at 3507, 3'-CCAGACC-5' at 3550, 3'-CTGTTCC-5' at 3625, 3'-TCACACC-5' at 3824, 3'-TCATTTT-5' at 4120, 3'-TCACTCT-5' at 4128, 3'-TTGATTT-5' at 4134, and 3'-TTAGTTT-5' at 4139.
These are 38 BBCABW Inrs on the negative strand in the positive direction: 3'-TTCACT-5', 2304, 3'-TGCAGT-5', 2328, 3'-GTCACT-5', 2425, 3'-GTCAGA-5', 2609, 3'-CTCAGA-5', 2699, 3'-TGCAGA-5', 2721, 3'-CTCAGA-5', 2729, 3'-TGCAGA-5', 2859, 3'-CTCAGA-5', 2866, 3'-CTCATT-5', 2902, 3'-GTCACT-5', 2929, 3'-TTCAGT-5', 2936, 3'-TGCACA-5', 2962, 3'-TGCATT-5', 3072, 3'-CCCAGT-5', 3082, 3'-CCCAGA-5', 3091, 3'-TCCACA-5', 3192, 3'-CTCACA-5', 3209, 3'-GCCAGA-5', 3221, 3'-TGCAGT-5', 3232, 3'-TGCAGT-5', 3281, 3'-CTCACT-5', 3317, 3'-TGCACT-5', 3343, 3'-CCCAGT-5', 3379, 3'-CCCACT-5', 3388, 3'-GGCACA-5', 3409, 3'-TGCAGT-5', 3461, 3'-GGCAGA-5', 3473, 3'-CTCACA-5', 3505, 3'-GCCACA-5', 3705, 3'-TCCAGA-5', 3806, 3'-GTCACA-5', 3822, 3'-TGCAGA-5', 3831, 3'-TCCAGA-5', 3891, 3'-CGCAGA-5', 3916, 3'-GTCACA-5', 3954, 3'-TGCAGT-5', 3962, 3'-GGCACT-5', 4006, 3'-TCCACT-5', 4013.
These are 26 YYANWYY Inrs on the positive strand in the positive direction: 3'-CCGCACT-5' at 2555, 3'-CCGGTCC-5' at 2574, 3'-TCAGTCT-5' at 2609, 3'-TCAGTTC-5' at 2615, 3'-TCAGTCC-5' at 2620, 3'-CTATATT-5' at 2662, 3'-TCAATCC-5' at 2668, 3'-TCGTTTT-5' at 2707, 3'-TCGATTC-5' at 2789, 3'-TTGCTCC-5' at 2806, 3'-CTAAACT-5' at 2871, 3'-CTGGTCC-5' at 2876, 3'-CCAGACT-5' at 2943, 3'-CCGGACC-5' at 2988, 3'-CCAGACC-5', 3021, 3'-TTATACC-5' at 3162, 3'-CTGGTTT-5' at 3175, 3'-TCGGTCT-5' at 3221, 3'-CTACTCC-5' at 3478, 3'-CCGATCC-5' at 3484, 3'-TCGATCC-5' at 3522, 3'-CTGGTCT-5' at 3548, 3'-TCACACT-5' at 3594, 3'-CCACTCC-5' at 3647, 3'-CCGGACC-5' at 3679, 3'-CCGGACC-5' at 3758, 3'-CTGGACC-5' at 3787, 3'-TCACTCC-5' at 3878, 3'-TCAGACT-5' at 3924, 3'-TCACACC-5' at 3966, 3'-CCACACT-5' at 3971, 3'-TTACTCC-5' at 4096, 3'-CTACTCC-5', 4102, and 3'-CTAAATC-5' at 4136.
These are 25 BBCABW Inrs on the positive strand in the positive direction: 3'-TCCACT-5', 2375, 3'-CGCAGT-5', 2423, 3'-GTCACA-5', 2464, 3'-CCCAGA-5', 2489, 3'-TTCACT-5', 2511, 3'-CGCACT-5', 2555, 3'-GTCAGT-5', 2607, 3'-CTCAGT-5', 2613, 3'-TTCAGT-5', 2618, 3'-TCCATA-5', 2642, 3'-TCCAGA-5', 3019, 3'-CTCAGA-5', 3187, 3'-TGCAGA-5', 3256, 3'-CTCACA-5', 3592, 3'-GCCAGA-5', 3608, 3'-CTCACT-5', 3712, 3'-TCCATT-5', 3731, 3'-TCCAGA-5', 3771, 3'-CCCAGT-5', 3820, 3'-GTCACT-5', 3843, 3'-CTCACT-5', 3876, 3'-TTCAGA-5', 3922, 3'-TCCACT-5', 3934, 3'-GTCACA-5', 3964, 3'-CGCAGA-5', 4056.
Transcribed initiator elements[edit | edit source]
A Google Scholar search using A1BG and "initiator element" produced 2 results (0.07 sec):
- Identification of a novel enhancer region 1.7 Mb downstream of the c-myc gene controlling its expression in hematopoietic stem and progenitor cells, "Sequence elements found common to many core promoters include the TATA element (TBP-binding site), Inr (initiator element), BRE (TFIIB-recognition site), DPE (downstream promoter element), DCE (downstream core element) and MTE (motif ten element)." and 2. Molecular evolution of hominoid seminal plasma genes.
A Google Scholar search using Inr and "AGC box" produced About 3 results (0.04 sec):
- Jasmonic acid-responsive elements, (JAREs), "TGACG, Auxin and/or salicylic acid; perhaps light regulation [48]. INR, YTCANTYY, Light-responsive [49]. AGC-box or GCC-box, AGCCGCC 2, Ethylene (=ethylene-inducible defense genes) [50]."[14]
A Google Scholar search using Inr and "ATA box" produced About 27 results (0.06 sec): but apparently none together.
A Google Scholar search using Inr and "C box" produced About 223 results (0.06 sec):
- "The transcriptional start sites (-14, -19, and -33 bp) themselves all begin within the repeated element CTCAXXCT, which is similar to, although not identical with, the consensus Inr element for the TdT gene family (3). This element may play an important role in transcriptional activation since three initiation sites of the LH receptor gene begin within this repeated Inr-like sequence. [...] C-box protein does not appear to be the TATA-binding TFIID transcriptional factor since it is not competed by an unlabeled TATA oligomer [...], and there are no TATA like sequences within the protein-binding region (-42 and -73 bp)."[15]
- Inr has alternate meanings such as international normalised ratio (INR), Insulin Receptor (InR) and InR (Synthetic Initiation Region).
A Google Scholar search using Inr and "D box" produced About 290 results (0.05 sec):
- "Within the GR expression plasmid phGR (D4X), three amino acids required for proper receptor dimerization within the dimerization box (D-box) were exchanged (Heck et al, 1994).", in: "Glucocorticoid-dependent transcriptional repression of the osteocalcin gene by competitive binding at the TATA box".
- "D-box, aspartic acid-rich region" in "TFII-I and USF (RBF-2) regulate ras/MAPK-responsive HIV-1 transcription in t cells".
- Each "upstream 3 kb melanopsin promoter region (sense and antisense strands) was investigated for the presence of putative d-box (consensus sequence RTTAYGTAAY [62] with a cutoff score value [0.75) and e-box (consensus sequences CACGTG [62] and CACGTT [62]) motifs known to be important in circadian regulation of rhythmic clock genes. [...] Once identified, the flanking region of the TSS was compared to the consensus initiator (INR) sequence (YYCANWYY), where A represents the TSS [32]." in "Functional diversity of melanopsins and their global expression in the teleost retina."
Google Scholar: Your search - Inr "CARE" -"care" - did not match any articles.
A Google Scholar search using Inr and "CArG box" produced About 71 results (0.05 sec):
- "mSlo gene can use perfect and imperfect Inrs for the initiation of transcription. [...] one consensus (CArG box, CC(A/T)6GG; -1751CCTTTAAAGG-1742)", in: "Regulation of Mouse Slo Gene Expression MULTIPLE PROMOTERS, TRANSCRIPTION START SITES, AND GENOMIC ACTION OF ESTROGEN".
A Google Scholar search using Inr and "CRE box" produced 4 results (0.04 sec):
- insulin resistance (INR),
- "cyclicAMP response element (CRE) [...] the V𝛅1 promoter appears to belong to a recently described class of promoters that contain the degenerate YAYTCYYY consensus sequence termed the transcriptional initiator (Inr) element (65). In this type of promoter, transcription initiation usually begins at the A present in the consensus sequence and, indeed, some of the minor transcriptional start sites of the V𝛅1 gene can be localized to this sequence [...]; however, the pyrimidine-rich Inr-like elements may be found distal to the initiation site (66), as in the case of the V𝛅1 promoter, where an Inr-like sequence is present 16 bp upstream of the transcriptional startsite.", in: "Multiple cis-acting elements are required for proper transcription of the mouse V delta 1 T cell receptor promoter".
Google Scholar: Your search - Inr and "E box" produced About 2,600 results (0.07 sec):
- "The transcription factor TFII-I has been shown to bind independently to two distinct promoter
elements, a pyrimidine-rich initiator (Inr) and a recognition site (E-box) for upstream stimulatory factor 1 (USF1), and to stimulate USF1 binding to both of these sites.", in: "Cloning of an Inr‐and E‐box‐binding protein, TFII‐I, that interacts physically and functionally with USF1".
Google Scholar: Your search - Inr and "BREu" produced About 829 results (0.04 sec):
- In: "Control of human gene expression: High abundance of divergent transcription in genes containing both INR and BRE elements in the core promoter".
Google Scholar: Your search - Inr and "HNF6" produced About 59 results (0.05 sec):
- "The binding sites of transcription factors, such as FOXO1, PPAR-RXR, STAT, IK1, HNF6 and
HNF3, were predicted on PI3KC2b promoter and … In addition, several common transcription initiation elements, such as BRE, INR, DCE and DRE [6], also existed in the core promoters", in: "Functional Analysis of Promoters from Three Subtypes of the PI3K Family and Their Roles in the Regulation of Lipid Metabolism by Insulin in Yellow Catfish …".
- international normalized ratio (INR).
Google Scholar: Your search - Inr and "HY box" produced 2 results (0.04 sec): nada.
Google Scholar: Your search - Inr and "MRE" produced About 4,290 results (0.09 sec):
- "Relative locations of the TATAAAA (elipse) and CCAAT sequences are shown, as are the Inr sequence, metal responsive element (MRE) and the putative Antioxidant Responsive Element (ARE)." in: "Identification of a putative antioxidant response element in the 5′-flanking region of the human γ-glutamylcysteine synthetase heavy subunit gene".
Google Scholar: Your search - Inr and "Pyrimidine box" produced About 21 results (0.08 sec):
- Not mentioned together for one gene promoter.
Google Scholar: Your search - Inr and "STAT" produced About 27,600 results (0.06 sec):
- International Normalized Ratio (INR).
Google Scholar: Your search - "Initiator element" and "STAT" produced About 494 results (0.08 sec):
- "Activation of caspase-1 gene expression requires IRF-1 and Stat-1 (24, 32). Analysis of the human caspase-1 promoter has shown an IRF-1 binding site, which overlaps with the initiator element (33, 34)." in: "Role of p73 in regulating human caspase-1 gene transcription induced by interferon-γ and cisplatin".
Google Scholar: Your search - Inr and "TATA box" produced About 10,300 results (0.06 sec):
- "The locations of the TATA box and Inr element are indicated to the right." in: "Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization".
Google Scholar: Your search - Inr and "TAT box" produced About 13 results (0.07 sec): nada.
Google Scholar: Your search - Inr and "W box" produced About 105 results (0.06 sec):
- "Box T is bound by general transcription factors and is defined by position relative to the TATA box and Inr and not by … contains three W box sequences (TTGAC, TTGACC, GTCA)" in: "DNase1 footprints suggest the involvement of at least three types of transcription factors in the regulation of α-Amy2/A by gibberellin".
Laboratory reports[edit | edit source]
Below is an outline for sections of a report, paper, manuscript, log book entry, or lab book entry. You may create your own, of course.
Initiator element transcription laboratory
by --Marshallsumter (discuss • contribs) 00:32, 3 July 2019 (UTC)
Abstract[edit | edit source]
Inrs are apparently fairly common in gene promoters. They are present in the promoters of A1BG, and could make transcription easier. The presence of an Inr may indicate a gene silencer especially when a CAAT box is present. Testing these promoters for an Inr and possible interactions with other transcription factors already found increases the likelihood of multiple transcription pathways. At least one Inr has been found between neighboring gene ZSCAN22 and A1BG. The TATA boxes earlier discovered in the distal promoter on this same side may be indicative of a second transcription start site.
Introduction[edit | edit source]
Many transcription factors (TFs) may occur upstream and occasionally downstream of the transcription start site (TSS), in this gene's promoter. The following have been examined so far: (1) AGC boxes (GCC boxes), (2) ATA boxes, (3) CAAT boxes, (4) C and D boxes, (5) CAREs (GA responsive complexes), (6) CArG boxes, (7) CENP-B boxes, (8) CGCG boxes, (9) CRE boxes, (10) DREB boxes, (11) EIF4E basal elements (4EBEs), (12) enhancer boxes (E boxes), (13) E2 boxes, (14) Factor II B recognition elements, (15) GAREs (GA responsive complexes), (16) G boxes, (17) GC boxes, (18) GLM boxes, (19) HNF6s, (20) HY boxes, (21) Metal responsive elements (MREs), (22) Motif ten elements (MTEs), (23) Pyrimidine boxes (GA responsive complexes), (24) STAT5s, (25) TACTAAC boxes, (26) TATA boxes, (27) TAT boxes (GA responsive complexes), (28) TATCCAC boxes, (29) W boxes (GA responsive complexes), (30) X boxes and (31) Y boxes.
But, no (3) CAAT box, (7) CENP-B box, (8) CGCG boxes are too close to ZSCAN22, (10) no DREB box, (11) EIF4E basal element, (13) E2 boxes, (15) GARE are too close to ZSCAN22, (16) no G box, (18) GLM box, (22) MTE, (25) TACTAAC box, (27) a TAT box, (28) TATCCAC box, (30) X box, or (31) Y box occur.
Interactions may occur with (1) an AGC (GCC) box, (2) an ATA box, (4) C boxes, a D box, but the other C-box and D-box have not been tested, (5) CAREs, (6) CArG boxes, (9) a CRE box, (12) enhancer boxes, (14) a BREu, (17) GC boxes, (19) HNF6s, (20) HY boxes, (21) an MRE, (23) pyrimidine boxes, (24) STAT5s, (26) TATA boxes outside the core promoter, or (29) W boxes.
Experiments[edit | edit source]
Regarding hypothesis 1: Initiator elements are not present in the promoter of A1BG.
The Basic programs (starting with SuccessablesInr.bas and SuccessablesInr2.bas) were written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including the extended number of nts from 958 to 4445, looking for Inrs, their possible complements and inverses, to test the hypothesis that either consensus sequence YYANWYY or BBCABW are not present in the promoter of A1BG.
Regarding hypothesis 2: A1BG is not transcribed by a TATA box.
As TATA boxes do occur in the promoters of A1BG but not in the core promoters, can these TATA boxes interact with Inrs to become alternative transcription start sites?
Results[edit | edit source]
Hypothesis 1[edit | edit source]
For the consensus sequence YYANWYY: there is the following Inr in the core promoter, negative strand, negative direction: 3'-TTACTCC-5' at 4557, which is past the expected TSS at 4460.
For the consensus sequence YYANWYY: there are four Inrs in the core promoter, positive strand, negative direction: 3'-CCACTCC-5' at 4425, 3'-CCACTT+1T-5' at 4461, where the expected TSS is at 4460 as indicated, 3'-TCACATT-5' at 4533, and 3'-TTAATTC-5' at 4542. The remaining three are presumably downstream.
For the consensus sequence BBCABW: there are no Inr in the core promoter, negative strand, negative direction.
For the consensus sequence BBCABW: there are five Inrs in the core promoter, positive strand, negative direction: 3'-TCCACT-5', 4423, 3'-CCCAGA-5', 4448, 3'-TCCACT-5', 4459, 3'-CCCACT-5', 4485, 3'-TTCACA-5', 4531. None of these occur at the expected TSS of 4460.
From the ZNF497 positive direction toward A1BG, for the consensus sequence YYANWYY: there is the following Inr in the core promoter, negative strand, positive direction: 3'-CTGCACC-5' at 4343, which again is downstream.
For the consensus sequence YYANWYY: there are two Inrs in the core promoter, positive strand, positive direction: 3'-CCACTCC-5' at 4401 and 3'-CCAGACC-5' at 4416, both of which are downstream.
For the consensus sequence BBCABW: there are five Inr in the core promoter, negative strand, positive direction: 3'-GTCAGT-5', 4271, 3'-CTCATT-5', 4309, 3'-TGCAGA-5', 4317, 3'-CCCAGA-5', 4330, 3'-CTCACT-5', 4338. None of these occur at the known TSS.
For the consensus sequence BBCABW: there are four Inrs in the core promoter, positive strand, positive direction: 3'-TCCAGT-5', 4269, 3'-CTCACT-5', 4350, 3'-CCCACT-5', 4399, 3'-CCCAGA-5', 4414. These are also away from the known TSS.
Hypothesis 2[edit | edit source]
"During productive infection, human cytomegalovirus (HCMV) UL44 transcription initiates at three distinct start sites that are differentially regulated. Two of the start sites, the distal and the proximal, are active at early times [...]. The UL44 early viral gene product is essential for viral DNA synthesis. [...] The UL44 early viral promoters have a canonical TATA sequence, “TATAA.”"[16]
For the positive strand in the negative direction looking for 3'-TATA-A/T-A-A/T-A/G-5', there's one 3'-TATATAAA-5' at 2874 nts, its complement and inverse complement.
Discussions[edit | edit source]
If initiator elements can occur at additional TSS locations, then A1BG can have multiple TSSs.
Hypothesis 1 discussion[edit | edit source]
A1BG may be transcribed by multiple TSSs where each Inr occurs, whether these Inrs are upstream or downstream from the expected TSSs. As no Inr occurs exactly at either TSS, then it could be stated that A1BG is not transcribed by an initiator element.
Hypothesis 2 discussion[edit | edit source]
If multiple TSSs occur even in the distal promoter or closer to the neighboring genes ZSCAN22 and ZNF497, then A1BG could be transcribed by a TATA box and/or an Inr.
Further, all of the transcription factors found so far in the promoters of A1BG could participate in assisting transcription except ATA box, D box, CARE, HY box, Pyrimidine box, and TAT box which apparently do not interact with an Inr based on the results found in the Transcribed initiator elements section.
Conclusions[edit | edit source]
A1BG may be transcribed by multiple TSSs where each Inr occurs, whether these Inrs are upstream or downstream from the expected TSSs. As no Inr occurs exactly at either TSS, then it could be stated that A1BG is not transcribed by an initiator element. If multiple TSSs occur even in the distal promoter or closer to the neighboring genes ZSCAN22 and ZNF497, then A1BG could be transcribed by a TATA box and/or an Inr.
The key to assisting the recovery of any astronaut now depends on which means of transcription best moderates the effects of microgravity or irradiation. This may require extensive molecular genetic testing.
Laboratory evaluations[edit | edit source]
To assess your example, including your justification, analysis and discussion, I will provide such an assessment of my example for comparison and consideration.
Evaluation
No wet chemistry experiments were performed to confirm that Gene ID: 1 may be transcribed from either side using transcription factors in the core, proximal or distal promoters. The NCBI Gene database is generalized, whereas individual human genome testing could demonstrate that A1BG is transcribed from either side using known transcription factors. Sufficient nucleotides have been added to the data sets for the ZNF497 side to confirm likely transcription of A1BG by these known transcription factors.
See also[edit | edit source]
References[edit | edit source]
- ↑ DR Liston, PJ Johnson (1999). "Analysis of a Ubiquitous Promoter Element in a Primitive Eukaryote: Early Evolution of the Initiator Element". Molecular and Cellular Biology. 19 (3): 2380–8. PMID 10022924. Unknown parameter
|month=ignored (help);|access-date=requires|url=(help) - ↑ C Yang, E Bolotin, T Jiang, FM Sladek, E Martinez (2007). "Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters". Gene. 389 (1): 52–65. doi:10.1016/j.gene.2006.09.029. PMC 1955227. PMID 17123746. Unknown parameter
|month=ignored (help) - ↑ JE Purdy, BJ Mann, LT Pho, WA Petri Jr (1994). "Transient transfection of the enteric parasite Entamoeba histolytica and expression of firefly luciferase". Proceedings of the National Academy of Science USA. 91 (15): 7099–103. PMID 8041752. Retrieved 2012-06-10. Unknown parameter
|month=ignored (help) - ↑ 4.0 4.1 Hualin Xi, Yong Yu, Yutao Fu, Jonathan Foley, Anason Halees, and Zhiping Weng (June 2007). "Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1". Genome Research. 17 (6): 798–806. doi:10.1101/gr.5754707. PMC 1891339. PMID 17567998.
- ↑ Jennifer E.F. Butler, James T. Kadonaga (October 15, 2002). "The RNA polymerase II core promoter: a key component in the regulation of gene expression". Genes & Development. 16 (20): 2583–292. doi:10.1101/gad.1026202. PMID 12381658.
- ↑ Stephen T. Smale and James T. Kadonaga (July 2003). "The RNA Polymerase II Core Promoter" (PDF). Annual Review of Biochemistry. 72 (1): 449–79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. Retrieved 2012-05-07.
- ↑ 7.0 7.1 7.2 Jennifer F. Kugel and James A. Goodrich (2017). "Finding the start site: redefining the human initiator element" (PDF). Genes & Development. 31 (1–2): 1. doi:10.1101/gad.295980.117. Retrieved 9 May 2019.
- ↑ Thomas Shafee and Rohan Lowe (2017). "Eukaryotic and prokaryotic gene structure" (PDF). WikiJournal of Medicine. 4 (1): 2. doi:10.15347/wjm/2017.002. Retrieved 2017-04-06. Unknown parameter
|month=ignored (help) - ↑ Koichi Takayama, Ken-ichirou Morohashi, Shin-ichlro Honda, Nobuyuki Hara and Tsuneo Omura (1994). "Contribution of Ad4BP, a Steroidogenic Cell-Specific Transcription Factor, to Regulation of the Human CYP11A and Bovine CYP11B Genes through Their Distal Promoters". The Journal of Biochemistry. 116 (1): 193–203. doi:10.1093/oxfordjournals.jbchem.a124493. Retrieved 2017-08-16. Unknown parameter
|month=ignored (help) - ↑ Michelle Craig Barton, Navid Madani, and Beverly M. Emerson (1997). "Distal enhancer regulation by promoter derepression in topologically constrained DNA in vitro". Proceedings of the National Academy of Sciences of the United States of America. 94 (14): 7257–62. Retrieved 2017-08-16. Unknown parameter
|month=ignored (help) - ↑ A Aoyama, T Tamura, K Mikoshiba (1990). "Regulation of brain-specific transcription of the mouse myelin basic protein gene: function of the NFI-binding site in the distal promoter". Biochemical and Biophysical Research Communications. 167 (2): 648–53. doi:10.1016/0006-291X(90)92074-A. Retrieved 2012-12-13. Unknown parameter
|month=ignored (help) - ↑ J Gao and L Tseng (1996). "Distal Sp3 binding sites in the hIGBP-1 gene promoter suppress transcriptional repression in decidualized human endometrial stromal cells: identification of a novel Sp3 form in decidual cells". Molecular Endocrinology. 10 (6): 613–21. doi:10.1210/me.10.6.613. Retrieved 2012-12-13. Unknown parameter
|month=ignored (help) - ↑ Peter Pasceri, Dylan Pannell, Xiumei Wu, and James Ellis (1998). "Full activity from human β-globin locus control region transgenes requires 5′ HS1, distal β-globin promoter, and 3′ β-globin sequences". Blood. 92 (2): 653–63. Retrieved 2012-12-13. Unknown parameter
|month=ignored (help) - ↑ Orlene Guerra-Peraza, Ha Thuy Nguyen, Peter Stamp, and Jörg Leipner (June 2009). "ZmCOI6.1, a novel, alternatively spliced maize gene, whose transcript level changes under abiotic stress". Plant Science. 176 (6): 783–791. doi:10.1016/j.plantsci.2009.03.004. Retrieved 6 July 2019.
- ↑ Chon Hwa Tsai-Morris, Xuanzhu Xie, Wei Wang, Ellen Buczko, and Maria L. Dufau (25 February 1993). "Promoter and Regulatory Regions of the Rat Luteinizing Hormone Receptor Gene" (PDF). The Journal of Biological Chemistry. 268 (6): 4447–4452. Retrieved 6 July 2019.
- ↑ Hiroki Isomura, Mark F. Stinski, Ayumi Kudoh, Takayuki Murata, Sanae Nakayama, Yoshitaka Sato, Satoko Iwahori, Tatsuya Tsurumi (5 December 2007). "Noncanonical TATA Sequence in the UL44 Late Promoter of Human Cytomegalovirus Is Required for the Accumulation of Late Viral Transcripts". Journal of Virology. 82 (4): 1638. doi:10.1128/JVI.01917-07. Retrieved 20 April 2019.
 KSF
KSF