Neuroimaging
 From Wikidoc - Reading time: 8 min
From Wikidoc - Reading time: 8 min
Overview[edit | edit source]
Neuroimaging includes the use of various techniques to either directly or indirectly image the structure, function/pharmacology of the brain. It is a relatively new discipline within medicine and neuroscience.
Neuroimaging falls into two broad categories: structural imaging and functional imaging. Structural imaging deals with the structure of the brain and the diagnosis of gross (large scale) intracranial disease (such as tumor), and injury. Functional imaging is used to diagnose metabolic diseases and lesions on a finer scale (such as Alzheimer's disease) and also for neurological and cognitive science research and building brain-computer interfaces. Functional imaging enables, for example, the processing of information by centers in the brain to be visualized directly. Such processing causes the involved area of the brain to increase metabolism and "light up" on the scan.
Types of brain imaging[edit | edit source]
CAT[edit | edit source]
Computed Tomography (CT) or Computed Axial Tomography (CAT) scanning uses a series of x-rays of the head taken from many different directions. Typically used for quickly viewing brain injuries, CT scanning uses a computer program that performs a numerical integral calculation (the inverse Radon transform) on the measured x-ray series to estimate how much of an x-ray beam is absorbed in a small volume of the brain. Typically the information is presented as cross sections of the brain [1]. In approximation, the more dense a material is, the whiter a volume of it will appear on the scan (just as in the more familiar "flat" X-rays). CT scans are primarily used for evaluating swelling from tissue damage in the brain and in assessment of ventricle size. Modern CT scanning can provide reasonably good images in a matter of minutes.
MRI[edit | edit source]
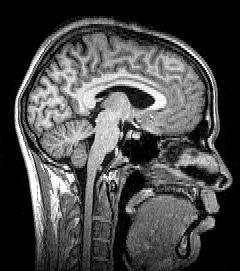
Magnetic Resonance Imaging (MRI) uses magnetic fields and radio waves to produce high quality two- or three-dimensional images of brain structures without use of ionizing radiation (X-rays) or radioactive tracers. During an MRI, a large cylindrical magnet creates a magnetic field around the head of the patient through which radio waves are sent. When the magnetic field is imposed, each point in space has a unique radio frequency at which the signal is received and transmitted (Preuss). Sensors read the frequencies and a computer uses the information to construct an image. The detection mechanisms are so precise that changes in structures over time can be detected. Using MRI, scientists can create images of both surface and subsurface structures with a high degree of anatomical detail. MRI scans can produce cross sectional images in any direction from top to bottom, side to side, or front to back. The problem with original MRI technology was that while it provides a detailed assessment of the physical appearance, water content, and many kinds of subtle derangements of structure of the brain (such as inflammation or bleeding), it fails to provide information about the metabolism of the brain (i.e. how actively it is functioning) at the time of imaging. A distinction is therefore made between "MRI imaging" and "functional MRI imaging" (fMRI), where MRI provides only structural information on the brain while fMRI yields both structural and functional data.
fMRI[edit | edit source]
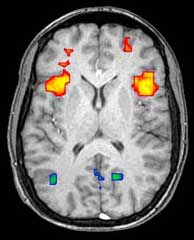
Functional Magnetic Resonance Imaging (fMRI) relies on the paramagnetic properties of oxygenated and deoxygenated hemoglobin to see images of changing blood flow in the brain associated with neural activity. This allows images to be generated that reflect which brain structures are activated (and how) during performance of different tasks. Most fMRI scanners allow subjects to be presented with different visual images, sounds and touch stimuli, and to make different actions such as pressing a button or moving a joystick. Consequently, fMRI can be used to reveal brain structures and processes associated with perception, thought and action. The resolution of fMRI is about 2-3 millimeters at present, limited by the spatial spread of the hemodynamic response to neural activity. It has largely superseded PET for the study of brain activation patterns. PET, however, retains the significant advantage of being able to identify specific brain receptors (or transporters) associated with particular neurotransmitters through its ability to image radiolabelled receptor "ligands" (receptor ligands are any chemicals that stick to receptors).
As well as research on healthy subjects, fMRI is increasingly used for the medical diagnosis of disease. Because fMRI is exquisitely sensitive to blood flow, it is extremely sensitive to early changes in the brain resulting from ischemia (abnormally low blood flow), such as the changes which follow stroke. Early diagnosis of certain types of stroke is increasingly important in neurology, since substances which dissolve blood clots may be used in the first few hours after certain types of stroke occur, but are dangerous to use afterwards. Brain changes seen on fMRI may help to make the decision to treat with these agents.
PET[edit | edit source]
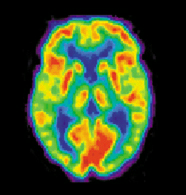
Positron Emission Tomography (PET) measures emissions from radioactively labeled metabolically active chemicals that have been injected into the bloodstream. The emission data are computer-processed to produce 2- or 3-dimensional images of the distribution of the chemicals throughout the brain (Nilsson 57). The positron emitting radioisotopes used are produced by a cyclotron, and chemicals are labelled with these radioactive atoms. The labeled compound, called a radiotracer, is injected into the bloodstream and eventually makes its way to the brain. Sensors in the PET scanner detect the radioactivity as the compound accumulates in various regions of the brain. A computer uses the data gathered by the sensors to create multicolored 2- or 3-dimensional images that show where the compound acts in the brain. Especially useful are a wide array of ligands used to map different aspects of neurotransmitter activity, with by far the most commonly used PET tracer being a labeled form of glucose (see FDG).
The greatest benefit of PET scanning is that different compounds can show blood flow and oxygen and glucose metabolism in the tissues of the working brain. These measurements reflect the amount of brain activity in the various regions of the brain and allow us to learn more about how the brain works. PET scans were superior to all other metabolic imaging methods in terms of resolution and speed of completion (as little as 30 seconds), when they first became available. The improved resolution permitted better study to be made as to the area of the brain activated by a particular task. The biggest drawback of PET scanning is that because the radioactivity decays rapidly, it is limited to monitoring short tasks (Nilsson 60). Before fMRI technology came online, PET scanning was the preferred method of functional (as opposed to structural) brain imaging, and it still continues to make large contributions to neuroscience.
PET scanning is also used for diagnosis of brain disease, most notably because brain tumors, strokes, and neuron-damaging diseases which cause dementia (such as Alzheimer's disease) all cause great changes in brain metabolism, which in turn causes easily detectable changes in PET scans. PET is probably most useful in early cases of certain dementias (with classic examples being Azheimer's disease and Pick's disease) where the early damage is too diffuse and makes too little difference in brain volume and gross structure to change CT and standard MRI images enough to be able to reliably differentiate it from the "normal" range of cortical atrophy which occurs with aging (in many but not all) persons, and which does not cause clinical dementia.
SPECT[edit | edit source]
Single Photon Emission Computed Tomography (SPECT) is similar to PET and uses gamma ray emitting radioisotopes and a gamma camera to record data that a computer uses to construct two- or three-dimensional images of active brain regions (Ball). SPECT relies on an injection of radioactive tracer, which is rapidly taken up by the brain but does not redistribute. Uptake of SPECT agent is nearly 100% complete within 30 – 60s, reflecting cerebral blood flow (CBF) at the time of injection. These properties of SPECT make it particularly well suited for epilepsy imaging, which is usually made difficult by problems with patient movement and variable seizure types. SPECT provides a "snapshot" of cerebral blood flow since scans can be acquired after seizure termination (so long as the radioactive tracer was injected at the time of the seizure). A significant limitation of SPECT is its poor resolution (about 1 cm) compared to that of MRI.
Like PET, SPECT also can be used to differentiate different kinds of disease process which produce dementia, and it is increasingly used for this purpose. Neuro-PET has a disadvantage of requiring use of a tracers with half-lives of at most 110 minutes, such as FDG. These must be made in a cyclotron, and are expensive or even unavailable if necessary transport times are prolonged more than a few half-lives. SPECT, however, is able to make use of tracers with much longer half-lives, such as technetium-99m, and as a result, is far more widely available.
DOT[edit | edit source]
Diffuse Optical Imaging (DOI) or Diffuse Optical Tomography (DOT) is a medical imaging modality which uses near infrared light to generate images of the body. The technique measures the optical absorption of haemoglobin, and relies on the absorption spectrum of haemoglobin varying with its oxygenation status.
History[edit | edit source]
In 1918 the American neurosurgeon Walter Dandy introduced the technique of ventriculography. X-ray images of the ventricular system within the brain were obtained by injection of filtered air directly into one or both lateral ventricles of the brain. Dandy also observed that air introduced into the subarachnoid space via lumbar spinal puncture could enter the cerebral ventricles and also demonstrate the cerebrospinal fluid compartments around the base of the brain and over its surface. This technique was called pneumoencephalography.
In 1927 Egas Moniz, professor of neurology in Lisbon, introduced cerebral angiography, whereby both normal and abnormal blood vessels in and around the brain could be visualized with great accuracy.
In the early 1970s, Allan McLeod Cormack and Godfrey Newbold Hounsfield introduced computerized axial tomography (CAT or CT scanning), and ever more detailed anatomic images of the brain became available for diagnostic and research purposes. Cormack and Hounsfield won the 1979 Nobel Prize for Physiology or Medicine for their work. Soon after the introduction of CAT in the early 1980s, the development of radioligands allowed single photon emission computed tomography (SPECT) and positron emission tomography (PET) of the brain.
More or less concurrently, magnetic resonance imaging (MRI or MR scanning) was developed by researchers including Peter Mansfield and Paul Lauterbur, who were awarded the Nobel Prize for Physiology or Medicine in 2003. In the early 1980s MRI was introduced clinically, and during the 1980s a veritable explosion of technical refinements and diagnostic MR applications took place. Scientists soon learned that the large blood flow changes measured by PET could also be imaged by the correct type of MRI. Functional magnetic resonance imaging (fMRI) was born, and since the 1990s, fMRI has come to dominate the brain mapping field due to its low invasiveness, lack of radiation exposure, and relatively wide availability. As noted above fMRI is also beginning to dominate the field of stroke treatment.
In early 2000s the field of neuroimaging reached the stage where limited practical applications of functional brain imaging have became feasible. The main application area is crude forms of brain-computer interface.
See also[edit | edit source]
- Functional neuroimaging
- functional near-infrared imaging
- History of brain imaging
- Human Cognome Project
- Medical imaging
- Brain mapping
- Statistical parametric mapping
- Neuroimaging software
- Voxel-based morphometry
References[edit | edit source]
- ↑ Jeeves, p. 21
Links[edit | edit source]
- The Whole Brain Atlas @ Harvard
- The American Society of Neuroimaging (ASN).
- UCLA Neuroimaging Training Program.
- Laboratory of Neuro Imaging at UCLA
Works cited[edit | edit source]
- Ball, Philip. "Brain Imaging Explained."
- Beaumont, J. Graham. Introduction to Neuropsychology. New York: The Guilford Press, 1983. 314 pages.
- Changeux, Jean-Pierre. Neuronal Man: The Biology of Mind. New York: Oxford University Press, 1985. 348 pages.
- Jeeves, Malcom. Mind Fields: Reflections on the Science of Mind and Brain. Grand Rapids, MI: Baker Books, 1994. 141 pages.
- Johnson, Keith A. "Neuroimaging Primer." [1]
- Leventon, Michael. "Transcranial Magnetic Stimulation." In association with MIT AI Lab. [2]
- Lister, Richard G. and Herbert J. Weingartner. Perspectives on Cognitive Neuroscience. New York: Oxford University Press, 1991. 508 pages.
- Mattson, James and Merrill Simon. The Pioneers of NMR and Magnetic Resonance in Medicine. United States: Dean Books Company, 1996. 838 pages.
- Nilsson, Lars-Goran and Hans J. Markowitsch. Cognitive Neuroscience of Memory. Seattle: Hogrefe & Huber Publishers, 1999. 307 pages.
- Norman, Donald A. Perspectives on Cognitive Science. New Jersey: Ablex Publishing Corporation, 1981. 303 pages.
- Pande, G.C. "Neurosciences and Philosophy." [3]
- Rapp, Brenda. The Handbook of Cognitive Neuropsychology. Ann Arbor, MI: Psychology Press, 2001. 652 pages.
- Shorey, Jamie. "Foundations of fMRI." [4]
Template:Psychiatry is:Heilaskönnun he:סריקת מוח sv:Neuroradiologi
Template:WH Template:WikiDoc Sources Template:Jb1 Template:Jb2
 KSF
KSF