Plasmin
 From Wikidoc - Reading time: 9 min
From Wikidoc - Reading time: 9 min
| VALUE_ERROR (nil) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Aliases | |||||||
| External IDs | GeneCards: [1] | ||||||
| Orthologs | |||||||
| Species | Human | Mouse | |||||
| Entrez |
|
| |||||
| Ensembl |
|
| |||||
| UniProt |
|
| |||||
| RefSeq (mRNA) |
|
| |||||
| RefSeq (protein) |
|
| |||||
| Location (UCSC) | n/a | n/a | |||||
| PubMed search | n/a | n/a | |||||
| Wikidata | |||||||
| |||||||
Plasmin is an important enzyme (EC 3.4.21.7) present in blood that degrades many blood plasma proteins, including fibrin clots. The degradation of fibrin is termed fibrinolysis. In humans, the plasmin protein is encoded by the PLG gene.[1]
Function[edit | edit source]
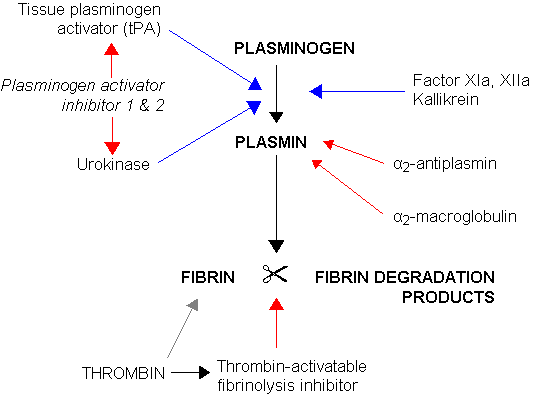
Plasmin is a serine protease that acts to dissolve fibrin blood clots. Apart from fibrinolysis, plasmin proteolyses proteins in various other systems: It activates collagenases, some mediators of the complement system, and weakens the wall of the Graafian follicle, leading to ovulation. It cleaves fibrin, fibronectin, thrombospondin, laminin, and von Willebrand factor. Plasmin, like trypsin, belongs to the family of serine proteases.
Plasmin is released as a zymogen called plasminogen (PLG) from the liver into the systemic circulation. Two major glycoforms of plasminogen are present in humans - type I plasminogen contains two glycosylation moieties (N-linked to N289 and O-linked to T346), whereas type II plasminogen contains only a single O-linked sugar (O-linked to T346). Type II plasminogen is preferentially recruited to the cell surface over the type I glycoform. Conversely, type I plasminogen appears more readily recruited to blood clots.
In circulation, plasminogen adopts a closed, activation resistant conformation. Upon binding to clots, or to the cell surface, plasminogen adopts an open form that can be converted into active plasmin by a variety of enzymes, including tissue plasminogen activator (tPA), urokinase plasminogen activator (uPA), kallikrein, and factor XII (Hageman factor). Fibrin is a cofactor for plasminogen activation by tissue plasminogen activator. Urokinase plasminogen activator receptor (uPAR) is a cofactor for plasminogen activation by urokinase plasminogen activator. The conversion of plasminogen to plasmin involves the cleavage of the peptide bond between Arg-561 and Val-562.[1][2][3][4]
Plasmin cleavage produces angiostatin.
Mechanism of plasminogen activation[edit | edit source]
Full length plasminogen comprises seven domains. In addition to a C-terminal chymotrypsin-like serine protease domain, plasminogen contains an N-terminal Pan Apple domain (PAp) together with five Kringle domains (KR1-5). The Pan-Apple domain contains important determinants for maintaining plasminogen in the closed form, and the kringle domains are responsible for binding to lysine residues present in receptors and substrates.
The X-ray crystal structure of closed plasminogen reveals that the PAp and SP domains maintain the closed conformation through interactions made throughout the kringle array .[4] Chloride ions further bridge the PAp / KR4 and SP / KR2 interfaces, explaining the physiological role of serum chloride in stabilizing the closed conformer. The structural studies also reveal that differences in glycosylation alter the position of KR3. These data help explain the functional differences between the type I and type II plasminogen glycoforms.
In closed plasminogen, access to the activation bond (R561/V562) targeted for cleavage by tPA and uPA is blocked through the position of the KR3/KR4 linker sequence and the O-linked sugar on T346. The position of KR3 may also hinder access to the activation loop. The Inter-domain interactions also block all kringle ligand-binding sites apart from that of KR-1, suggesting that the latter domain governs pro-enzyme recruitment to targets. Analysis of an intermediate plasminogen structure suggests that plasminogen conformational change to the open form is initiated through KR-5 transiently peeling away from the PAp domain. These movements expose the KR5 lysine-binding site to potential binding partners, and suggest a requirement for spatially distinct lysine residues in eliciting plasminogen recruitment and conformational change respectively.[4]
Mechanism of plasmin inactivation[edit | edit source]
This section does not cite any sources. (November 2016) (Learn how and when to remove this template message) |
Plasmin is inactivated by proteins such as α2-macroglobulin and α2-antiplasmin. The mechanism of plasmin inactivation involves the cleavage of an α2-macroglobulin at the bait region (a segment of the aM that is particularly susceptible to proteolytic cleavage) by plasmin. This initiates a conformational change such that the α2-macroglobulin collapses about the plasmin. In the resulting α2-macroglobulin-plasmin complex, the active site of plasmin is sterically shielded, thus substantially decreasing the plasmin's access to protein substrates. Two additional events occur as a consequence of bait region cleavage, namely (i) a h-cysteinyl-g-glutamyl thiol ester of the α2-macroglobulin becomes highly reactive and (ii) a major conformational change exposes a conserved COOH-terminal receptor binding domain. The exposure of this receptor binding domain allows the α2-macroglobulin protease complex to bind to clearance receptors and be removed from circulation.
Pathology[edit | edit source]
Plasmin deficiency may lead to thrombosis, as clots are not adequately degraded. Plasminogen deficiency in mice leads to defective liver repair,[5] defective wound healing, reproductive abnormalities.[citation needed]
In humans, a rare disorder called plasminogen deficiency type I (Online Mendelian Inheritance in Man (OMIM) 217090) is caused by mutations of the PLG gene and is often manifested by ligneous conjunctivitis.
Interactions[edit | edit source]
Plasmin has been shown to interact with Thrombospondin 1,[6][7] Alpha 2-antiplasmin[8][9] and IGFBP3.[10] Moreover, plasmin induces the generation of bradykinin in mice and humans through high molecular weight kininogen cleavage.[11]
References[edit | edit source]
- ↑ 1.0 1.1 "Entrez Gene: plasminogen".
- ↑ Miyata T, Iwanaga S, Sakata Y, Aoki N (October 1982). "Plasminogen Tochigi: inactive plasmin resulting from replacement of alanine-600 by threonine in the active site". Proc. Natl. Acad. Sci. U.S.A. 79 (20): 6132–6. Bibcode:1982PNAS...79.6132M. doi:10.1073/pnas.79.20.6132. PMC 347073. PMID 6216475.
- ↑ Forsgren M, Råden B, Israelsson M, Larsson K, Hedén LO (March 1987). "Molecular cloning and characterization of a full-length cDNA clone for human plasminogen". FEBS Lett. 213 (2): 254–60. doi:10.1016/0014-5793(87)81501-6. PMID 3030813.
- ↑ 4.0 4.1 4.2 Law RH, Caradoc-Davies T, Cowieson N, Horvath AJ, Quek AJ, Encarnacao JA, Steer D, Cowan A, Zhang Q, Lu BG, Pike RN, Smith AI, Coughlin PB, Whisstock JC (2012). "The X-ray crystal structure of full-length human plasminogen". Cell Rep. 1 (3): 185–90. doi:10.1016/j.celrep.2012.02.012. PMID 22832192.
- ↑ Bezerra JA, Bugge TH, Melin-Aldana H, Sabla G, Kombrinck KW, Witte DP, Degen JL (December 21, 1999). "Plasminogen deficiency leads to impaired remodeling after a toxic injury to the liver". Proc. Natl. Acad. Sci. U.S.A. Proceedings of the National Academy of Sciences of the United States of America. 96 (26): 15143–8. Bibcode:1999PNAS...9615143B. doi:10.1073/pnas.96.26.15143. PMC 24787. PMID 10611352.
- ↑ Silverstein RL, Leung LL, Harpel PC, Nachman RL (November 1984). "Complex formation of platelet thrombospondin with plasminogen. Modulation of activation by tissue activator". J. Clin. Invest. 74 (5): 1625–33. doi:10.1172/JCI111578. PMC 425339. PMID 6438154.
- ↑ DePoli P, Bacon-Baguley T, Kendra-Franczak S, Cederholm MT, Walz DA (March 1989). "Thrombospondin interaction with plasminogen. Evidence for binding to a specific region of the kringle structure of plasminogen". Blood. 73 (4): 976–82. PMID 2522013.
- ↑ Wiman B, Collen D (September 1979). "On the mechanism of the reaction between human alpha 2-antiplasmin and plasmin". J. Biol. Chem. 254 (18): 9291–7. PMID 158022.
- ↑ Shieh BH, Travis J (May 1987). "The reactive site of human alpha 2-antiplasmin". J. Biol. Chem. 262 (13): 6055–9. PMID 2437112.
- ↑ Campbell PG, Durham SK, Suwanichkul A, Hayes JD, Powell DR (August 1998). "Plasminogen binds the heparin-binding domain of insulin-like growth factor-binding protein-3". Am. J. Physiol. 275 (2 Pt 1): E321–31. PMID 9688635.
- ↑ Marcos-Contreras OA, Martinez de Lizarrondo S, Bardou I, Orset C, Pruvost M, Anfray A, Frigout Y, Hommet Y, Lebouvier L, Montaner J, Vivien D, Gauberti M (August 2016). "Hyperfibrinolysis increases blood brain barrier permeability by a plasmin and bradykinin-dependent mechanism". Blood. 128: 2423–2434. doi:10.1182/blood-2016-03-705384. PMID 27531677.
Further reading[edit | edit source]
- Shanmukhappa K, Mourya R, Sabla GE, Degen JL, Bezerra JA (July 2005). "Hepatic to pancreatic switch defines a role for hemostatic factors in cellular plasticity in mice". Proc. Natl. Acad. Sci. U.S.A. 102 (29): 10182–7. Bibcode:2005PNAS..10210182S. doi:10.1073/pnas.0501691102. PMC 1177369. PMID 16006527.
- Anglés-Cano E, Rojas G (2002). "Apolipoprotein(a): structure-function relationship at the lysine-binding site and plasminogen activator cleavage site". Biol. Chem. 383 (1): 93–9. doi:10.1515/BC.2002.009. PMID 11928826.
- Ranson M, Andronicos NM (2003). "Plasminogen binding and cancer: promises and pitfalls". Front. Biosci. 8: s294–304. doi:10.2741/1044. PMID 12700073.
External links[edit | edit source]
- The MEROPS online database for peptidases and their inhibitors: S01.233
- Plasmin at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
Lua error in Module:Authority_control at line 788: attempt to index field 'wikibase' (a nil value).
 KSF
KSF