Rossmann fold
 From Wikidoc - Reading time: 2 min
From Wikidoc - Reading time: 2 min
Overview[edit | edit source]
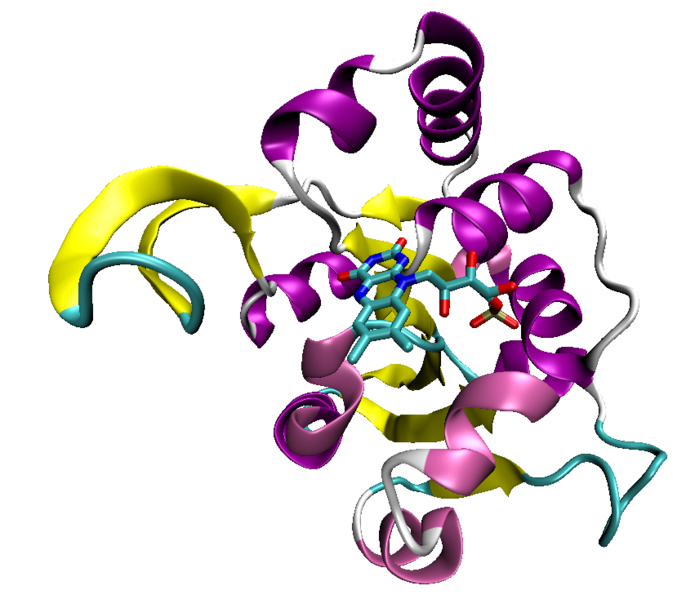
The Rossmann fold is a protein structural motif found in proteins that bind nucleotides, especially the cofactor NAD. The structure is composed of three or more parallel beta strands linked by two alpha helices in the topological order beta-alpha-beta-alpha-beta. Because each Rossmann fold can bind one nucleotide, binding domains for dinucleotides such as NAD consist of two paired Rossmann folds that each bind one nucleotide moiety of the cofactor molecule. Single Rossmann folds can bind mononucleotides such as the cofactor FMN.
The motif is named for Michael Rossmann who first pointed out that this was a frequently occurring motif in nucleotide binding proteins.[1]
References[edit | edit source]
- ↑ Rao S, Rossmann M (1973). "Comparison of super-secondary structures in proteins". J Mol Biol. 76 (2): 241–56. doi:10.1016/0022-2836(73)90388-4. PMID 4737475.
Licensed under CC BY-SA 3.0 | Source: https://www.wikidoc.org/index.php/Rossmann_fold18 views | Status: cached on January 15 2026 20:04:40↧ Download this article as ZWI file
 KSF
KSF