STAT5
 From Wikidoc - Reading time: 8 min
From Wikidoc - Reading time: 8 min
| signal transducer and activator of transcription 5A | |
|---|---|
| File:Protein STAT5A PDB 1y1u.png STAT5A | |
| Identifiers | |
| Symbol | STAT5A |
| Alt. symbols | STAT5 |
| Entrez | 6776 |
| HUGO | 11366 |
| OMIM | 601511 |
| RefSeq | NM_003152 |
| UniProt | P42229 |
| Other data | |
| Locus | Chr. 17 q11.2 |
| signal transducer and activator of transcription 5B | |
|---|---|
| File:Protein STAT5B PDB 1y1u.png STAT5B | |
| Identifiers | |
| Symbol | STAT5B |
| Entrez | 6777 |
| HUGO | 11367 |
| OMIM | 604260 |
| RefSeq | NM_012448 |
| UniProt | P51692 |
| Other data | |
| Locus | Chr. 17 q11.2 |
Signal transducer and activator of transcription 5 (STAT5) refers to two highly related proteins, STAT5A and STAT5B, which are part of the seven-membered STAT family of proteins. Though STAT5A and STAT5B are encoded by separate genes, the proteins are 90% identical at the amino acid level.[1] STAT5 proteins are involved in cytosolic signalling and in mediating the expression of specific genes.[2] Aberrant STAT5 activity has been shown to be closely connected to a wide range of human cancers,[3] and silencing this aberrant activity is an area of active research in medicinal chemistry.[4]
Activation and function[edit | edit source]
In order to be functional, STAT5 proteins must first be activated. This activation is carried out by kinases associated with transmembrane receptors:[3]
- first, ligands binding to these transmembrane receptors on the outside of the cell activate the kinases;
- second, the stimulated kinases add a phosphate group to a specific tyrosine residue on the receptor;
- STAT5 then binds to these phosphorylated-tyrosines using their SH2 domain (STAT domains illustrated below);
- the bound STAT5 is then phosphorylated by the kinase, the phosphorylation occurring at particular tyrosine residues on the C-terminus of the protein;
- phosphorylation causes STAT5 to dissociate from the receptor;
- the phosphorylated STAT5 finally goes on to form either homodimers, STAT5-STAT5, or heterodimers, STAT5-STATX, with other STAT proteins. The SH2 domains of the STAT5 proteins are once again used for this dimerization. STAT5 can also form homo-tetramers, usually in concert with the histone methyltransferase EZH2, and act as a transcriptional repressor.[5]
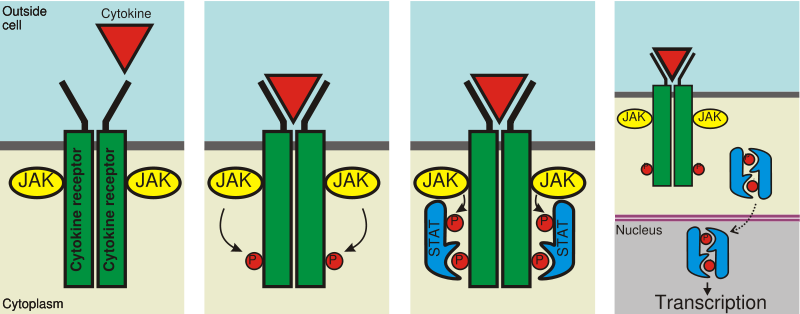
In the activation pathway illustrated to the left, the ligand involved is a cytokine and the specific kinase taking part in activation is JAK. The dimerized STAT5 represents the active form of the protein, which is ready for translocation into the nucleus.
Once in the nucleus, the dimers bind to STAT5 response elements, inducing transcription of specific sets of genes. Upregulation of gene expression by STAT5 dimers has been observed for genes dealing with:[2]
- controlled cell growth and division, or cell proliferation
- programmed cell death, or apoptosis
- cell specialization, or differentiation and
- inflammation.
Activated STAT5 dimers are, however, short-lived and the dimers are made to undergo rapid deactivation. Deactivation may be carried out by a direct pathway, removing the phosphate groups using phosphatases like PIAS or SHP-2 for example, or by an indirect pathway, which involves reducing cytokine signalling.[6]
STAT5 and cancer[edit | edit source]
STAT5 has been found to be constitutively phosphorylated in cancer cells,[4] implying that the protein is always present in its active form. This constant activation is brought about either by mutations or by aberrant expressions of cell signalling, resulting in poor regulation, or complete lack of control, of the activation of transcription for genes influenced by STAT5. This leads to constant and increased expression of these genes. For example, mutations may lead to increased expression of anti-apoptotic genes, the products of which actively prevent cell death. The constant presence of these products preserve the cell in spite of it having become cancerous, causing the cell to eventually become malignant.
Treatment approaches[edit | edit source]
Attempts at treatment for cancer cells with constitutively phosphorylated STAT5 have included both indirect and direct inhibition of STAT5 activity. While more medicinal work has been done in indirect inhibition, this approach can lead to increased toxicity in cells and can also result in non-specific effects, both of which are better handled by direct inhibition.[4]
Indirect inhibition targets kinases associated with STAT5, or targets proteases that carry out terminal truncation of proteins. Different inhibitors have been designed to target different kinases:
- inhibition of BCR/ABl constitutes the basis of the functioning of drugs like imatinib[7]
- inhibition of FLT3 is carried out by drugs like lestaurtinib[8]
- inhibition of JAK2 is carried out by the drug CYT387, which was successful in preclinical trials and is currently undergoing clinical trials.[9]
Direct inhibition of STAT5 activity makes use of small molecule inhibitors that prevent STAT5 from properly binding to DNA, or prevent proper dimerization. The inhibiting of DNA binding utilizes RNA interference,[10] antisense oligodeoxynucleotide,[10] and short hairpin RNA.[11] The inhibition of proper dimerization, on the other hand, is brought about by the use of small molecules that target the SH2 domain. Recent work on drug development in the latter field have proved particularly effective.[12]
References[edit | edit source]
- ↑ Grimley PM, Dong F, Rui H (June 1999). "Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation". Cytokine Growth Factor Rev. 10 (2): 131–57. doi:10.1016/S1359-6101(99)00011-8. PMID 10743504.
- ↑ 2.0 2.1 Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T (September 1999). "STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells". EMBO J. 18 (17): 4754–65. doi:10.1093/emboj/18.17.4754. PMC 1171548. PMID 10469654.
- ↑ 3.0 3.1 Tan SH, Nevalainen MT (June 2008). "Signal transducer and activator of transcription 5A/B in prostate and breast cancers" (PDF). Endocr. Relat. Cancer. 15 (2): 367–90. doi:10.1677/ERC-08-0013. PMID 18508994.
- ↑ 4.0 4.1 4.2 Cumaraswamy AA, Todic A, Resetca D, Minden MD, Gunning PT (January 2012). "Inhibitors of Stat5 protein signalling". MedChemComm. 3 (1): 22. doi:10.1039/C1MD00175B.
- ↑ Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, Dinner AR, Clark MR (December 2011). "Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2". Nat. Immunol. 12 (12): 1212–20. doi:10.1038/ni.2136. PMC 3233979. PMID 22037603.
- ↑ Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers CL (July 1996). "Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia". Oncogene. 13 (2): 247–54. PMID 8710363.
- ↑ Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB (May 1996). "Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells". Nat. Med. 2 (5): 561–6. doi:10.1038/nm0596-561. PMID 8616716.
- ↑ Levis M, Allebach J, Tse KF, Zheng R, Baldwin BR, Smith BD, Jones-Bolin S, Ruggeri B, Dionne C, Small D (June 2002). "A FLT3-targeted tyrosine kinase inhibitor is cytotoxic to leukemia cells in vitro and in vivo". Blood. 99 (11): 3885–91. doi:10.1182/blood.V99.11.3885. PMID 12010785.
- ↑ Pardanani A, Lasho T, Smith G, Burns CJ, Fantino E, Tefferi A (August 2009). "CYT387, a selective JAK1/JAK2 inhibitor: in vitro assessment of kinase selectivity and preclinical studies using cell lines and primary cells from polycythemia vera patients". Leukemia. 23 (8): 1441–5. doi:10.1038/leu.2009.50. PMID 19295546.
- ↑ 10.0 10.1 Behbod F, Nagy ZS, Stepkowski SM, Karras J, Johnson CR, Jarvis WD, Kirken RA (October 2003). "Specific inhibition of Stat5a/b promotes apoptosis of IL-2-responsive primary and tumor-derived lymphoid cells" (PDF). J. Immunol. 171 (8): 3919–27. doi:10.4049/jimmunol.171.8.3919. PMID 14530308.
- ↑ Klosek SK, Nakashiro K, Hara S, Goda H, Hamakawa H (October 2008). "Stat3 as a molecular target in RNA interference-based treatment of oral squamous cell carcinoma". Oncol. Rep. 20 (4): 873–8. doi:10.3892/or_00000085. PMID 18813829.
- ↑ Page BD, Khoury H, Laister RC, Fletcher S, Vellozo M, Manzoli A, Yue P, Turkson J, Minden MD, Gunning PT (February 2012). "Small molecule STAT5-SH2 domain inhibitors exhibit potent antileukemia activity". J. Med. Chem. 55 (3): 1047–55. doi:10.1021/jm200720n. PMID 22148584.
 KSF
KSF