Divisome
 From Wikipedia - Reading time: 6 min
From Wikipedia - Reading time: 6 min
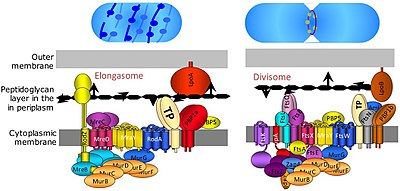
The divisome is a protein complex in bacteria that is responsible for cell division, constriction of inner and outer membranes during division, and peptidoglycan (PG) synthesis at the division site. The divisome is a membrane protein complex with proteins on both sides of the cytoplasmic membrane. In gram-negative cells it is located in the inner membrane. The divisome is nearly ubiquitous in bacteria although its composition may vary between species.[2]
The elongasome is a modified version of the divisome, without the membrane-constricting FtsZ-ring and its associated machinery. The elongasome is present only in non-spherical bacteria and directs lateral insertion of PG along the long axis of the cell, thus allowing cylindrical growth (as opposed to spherical growth, as in cocci).[2]
History
[edit]Some of the first cell-division genes of Escherichia coli were discovered by François Jacob's group in France in the 1960s. They were called fts genes, because mutants of these genes conferred a filamentous temperature-sensitive phenotype.[3] At the non-permissive temperature (usually 42 °C), fts mutant cells continue to elongate without dividing, forming filaments that can be up to 150 m long (as opposed to 2-3 m in wild-type cells). Three breakthroughs came with the discovery of the ftsZ gene in 1980[4] and the realization that the FtsZ protein was localized to the division plane of dividing cells,[5] and finally the realization that the structure of FtsZ is remarkably similar to tubulin and that they likely share a common ancestor.[6]
Composition
[edit]The precise composition of the divisome and elongasome remains unknown, given that they are highly dynamic protein complexes which recruit and release certain proteins during cell division. However, more than 20 proteins are known to be part of the divisome in E. coli with a similar number of proteins in Gram-positive bacteria (such as Bacillus subtilis), although not all proteins are conserved across bacteria.[7]
Several other fts genes, such as ftsA, ftsW, ftsQ, ftsI, ftsL, ftsK, ftsN, and ftsB, were all found to be essential for cell division and to associate with the divisome complex and the FtsZ ring. FtsA protein binds directly to FtsZ in the cytoplasm, and FtsB, FtsL and FtsQ form an essential membrane-embedded subcomplex. FtsK and FtsW are larger proteins with multiple transmembrane domains. FtsI, also known as PBP3, is the divisome-specific transpeptidase required for synthesis of the division septum.
DNA replication and cell division
[edit]DNA replication in bacteria is tightly linked to cell division. For instance, blocking replication in B. subtilis results in elongated cells without proper cell division. Bacterial DNA replication is initiated by the binding of DnaA (an ATPase) to the origin of replication (oriC) at midcell. FtsZ assembly appears to be linked to successful DNA replication[7] with MatP and ZapB somehow coordinating interactions between the division machinery and DNA replication during chromosome segregation in E. coli.[8]
Assembly of the divisome
[edit]The precise assembly process of the divisome is not well understood. It starts with the early proteins FtsZ and its membrane anchor FtsA, and the proteins ZipA, EzrA, and the Zaps (ZapA, ZapB, ZapC, ZapD) which promote FtsZ ring-formation. While FtsA and especially FtsZ are highly conserved among bacteria, ZipA, which is a second membrane anchor for FtsZ in gamma-proteobacteria, EzrA, and the Zap proteins are less well conserved and are missing in some species.[7] After the early proteins, the FtsQLB subcomplex is added,[9] followed by FtsI (transpeptidase), FtsW (transglycosylase), and FtsN. Both FtsI and FtsW are required for synthesis of the septal wall.[10] FtsW is related to the putative elongation-specific transglycosylase RodA, another divisome protein.[11] FtsN appears to have several functions: it stabilizes the divisome (at least when over-expressed), acts as a trigger for cytokinesis (via interactions with FtsI and FtsW), and activates FtsA mediated recruitment of FtsQLB[12][13] through direct binding of FtsA.[14] However, while FtsA, FtsQLB, FtsI and FtsW are widely conserved, FtsN is limited to Gram-negative organisms (such as E. coli) and hence is not universally required.[7]
The mitochondrial (eukaryotic) divisome
[edit]A protein complex that orchestrates division of eukaryotic mitochondria has been called the "mitochondrial divisome". It is conceptually and operationally similar to the bacterial cell-division machinery but consists (mostly) of different proteins. However, there seem to be some conserved aspects, e.g. in the red alga Cyanidioschyzon merolae, a mitochondrial FtsZ protein partially constricts the organelle, which enables the dynamin homologue Dnm1 to assemble with the mitochondrion-dividing (MD) ring on the cytosolic face to induce fission. However, in many eukaryotes (including yeasts and animals), the divisome functions in the complete absence of the contractile FtsZ ring.[15]
See also
[edit]- Fission (biology) – Biological process
References
[edit]- ^ Hugonnet, Jean-Emmanuel; Mengin-Lecreulx, Dominique; Monton, Alejandro; Blaauwen, Tanneke den; Carbonnelle, Etienne; Veckerlé, Carole; Yves; Brun, V.; Nieuwenhze, Michael van (2016-10-21). "Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli". eLife. 5. doi:10.7554/elife.19469. ISSN 2050-084X. PMC 5089857. PMID 27767957.
- ^ a b Szwedziak, Piotr; Löwe, Jan (December 2013). "Do the divisome and elongasome share a common evolutionary past?". Current Opinion in Microbiology. 16 (6): 745–751. doi:10.1016/j.mib.2013.09.003. ISSN 1879-0364. PMID 24094808.
- ^ Hirota, Y.; Ryter, A.; Jacob, F. (1968). "Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division". Cold Spring Harbor Symposia on Quantitative Biology. 33: 677–693. doi:10.1101/sqb.1968.033.01.077. ISSN 0091-7451. PMID 4892005.
- ^ Lutkenhaus, J. F.; Wolf-Watz, H.; Donachie, W. D. (May 1980). "Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ)". Journal of Bacteriology. 142 (2): 615–620. doi:10.1128/JB.142.2.615-620.1980. ISSN 0021-9193. PMC 294035. PMID 6991482.
- ^ Bi, E. F.; Lutkenhaus, J. (1991-11-14). "FtsZ ring structure associated with division in Escherichia coli". Nature. 354 (6349): 161–164. Bibcode:1991Natur.354..161B. doi:10.1038/354161a0. ISSN 0028-0836. PMID 1944597. S2CID 4329947.
- ^ Löwe, J.; Amos, L. A. (1998-01-08). "Crystal structure of the bacterial cell-division protein FtsZ". Nature. 391 (6663): 203–206. Bibcode:1998Natur.391..203L. doi:10.1038/34472. ISSN 0028-0836. PMID 9428770. S2CID 4330857.
- ^ a b c d den Blaauwen, Tanneke; Hamoen, Leendert W; Levin, Petra Anne (2017-04-01). "The divisome at 25: the road ahead". Current Opinion in Microbiology. Cell regulation. 36: 85–94. doi:10.1016/j.mib.2017.01.007. ISSN 1369-5274. PMC 6436919. PMID 28254403.
- ^ Espéli, Olivier; Borne, Romain; Dupaigne, Pauline; Thiel, Axel; Gigant, Emmanuelle; Mercier, Romain; Boccard, Frédéric (2012-07-18). "A MatP–divisome interaction coordinates chromosome segregation with cell division in E. coli". The EMBO Journal. 31 (14): 3198–3211. doi:10.1038/emboj.2012.128. ISSN 0261-4189. PMC 3400007. PMID 22580828.
- ^ Buddelmeijer, Nienke; Beckwith, Jon (June 2004). "A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region". Molecular Microbiology. 52 (5): 1315–1327. doi:10.1111/j.1365-2958.2004.04044.x. ISSN 0950-382X. PMID 15165235.
- ^ Wang, Lilin; Khattar, Medhat K.; Donachie, W. D.; Lutkenhaus, Joe (1998-06-01). "FtsI and FtsW Are Localized to the Septum inEscherichia coli". Journal of Bacteriology. 180 (11): 2810–2816. doi:10.1128/JB.180.11.2810-2816.1998. ISSN 1098-5530. PMC 107242. PMID 9603865.
- ^ Meeske, Alexander J.; Riley, Eammon P.; Robins, William P.; Uehara, Tsuyoshi; Mekalanos, John J.; Kahne, Daniel; Walker, Suzanne; Kruse, Andrew C.; Bernhardt, Thomas G.; Rudner, David Z. (September 2016). "SEDS proteins are a widespread family of bacterial cell wall polymerases". Nature. 537 (7622): 634–638. Bibcode:2016Natur.537..634M. doi:10.1038/nature19331. ISSN 1476-4687. PMC 5161649. PMID 27525505.
- ^ Dai, K; Xu, Y; Lutkenhaus, J (1993). "Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts)". Journal of Bacteriology. 175 (12): 3790–3797. doi:10.1128/JB.175.12.3790-3797.1993. ISSN 0021-9193. PMC 204796. PMID 8509333.
- ^ Pichoff, Sebastien; Du, Shishen; Lutkenhaus, Joe (2015). "The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA–FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring". Molecular Microbiology. 95 (6): 971–987. doi:10.1111/mmi.12907. ISSN 1365-2958. PMC 4364298. PMID 25496259.
- ^ Busiek, Kimberly K; Eraso, Jesus M; Wang, Yipeng; Margolin, William (2012). "The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN". Journal of Bacteriology. 194 (8): 1989–2000. doi:10.1128/JB.06683-11. PMC 3318488. PMID 22328664.
- ^ Kraus, Felix; Roy, Krishnendu; Pucadyil, Thomas J.; Ryan, Michael T. (February 2021). "Function and regulation of the divisome for mitochondrial fission". Nature. 590 (7844): 57–66. doi:10.1038/s41586-021-03214-x. ISSN 1476-4687. PMID 33536648. S2CID 231805756.
 KSF
KSF