Influenza A virus
 From Wikipedia - Reading time: 26 min
From Wikipedia - Reading time: 26 min
| Alphainfluenzavirus influenzae | |
|---|---|
| |
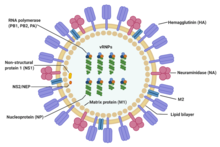
| Structure of influenza A virus | |
| |
| TEM micrograph of influenza A viruses | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Insthoviricetes |
| Order: | Articulavirales |
| Family: | Orthomyxoviridae |
| Genus: | Alphainfluenzavirus |
| Species: | Alphainfluenzavirus influenzae
|
Influenza A virus (Alphainfluenzavirus influenzae)[1] or IAV is the only species of the genus Alphainfluenzavirus of the virus family Orthomyxoviridae.[2] It is a pathogen with strains that infect birds and some mammals, as well as causing seasonal flu in humans.[3] Mammals in which different strains of IAV circulate with sustained transmission are bats, pigs, horses and dogs; other mammals can occasionally become infected.[4][5]
IAV is an enveloped negative-sense RNA virus, with a segmented genome.[5] Through a combination of mutation and genetic reassortment the virus can evolve to acquire new characteristics, enabling it to evade host immunity and occasionally to jump from one species of host to another.[6][7]
Subtypes of IAV are defined by the combination of the antigenic H and N proteins in the viral envelope; for example, "H1N1" designates an IAV subtype that has a type-1 hemagglutinin (H) protein and a type-1 neuraminidase (N) protein.[8] Almost all possible combinations of H (1 through 16) and N (1 through 11) have been isolated from wild birds.[9] Further variations exist within the subtypes and can lead to very significant differences in the virus's ability to infect and cause disease, as well as to the severity of symptoms.[10][11]
Symptoms of human seasonal flu usually include fever, cough, sore throat, muscle aches, conjunctivitis and, in severe cases, breathing problems and pneumonia that may be fatal.[12][3] Humans can rarely become infected with strains of avian or swine influenza, usually as a result of close contact with infected animals; symptoms range from mild to severe including death.[13][14] Bird-adapted strains of the virus can be asymptomatic in some aquatic birds but lethal if they spread to other species, such as chickens.[15]
IAV disease in poultry can be prevented by vaccination; however, biosecurity control measures are preferred.[16][17] In humans, seasonal influenza can be treated in its early stages with antiviral medicines.[18] A global network, the Global Influenza Surveillance and Response System (GISRS) monitors the spread of influenza with the aim to inform development of both seasonal and pandemic vaccines.[19] Several millions of specimens are tested by the GISRS network annually through a network of laboratories in 127 countries. As well as human viruses, GISRS monitors avian, swine, and other potentially zoonotic influenza viruses. IAV vaccines need to be reformulated regularly in order to keep up with changes in the virus.[20]
Virology
[edit]Classification
[edit]There are two methods of classification, one based on surface proteins (originally serotypes),[21] and the other based on its behavior, mainly the host animal.
Subtypes
[edit]
There are two antigenic proteins on the surface of the viral envelope, hemagglutinin and neuraminidase.[22] Different influenza virus genomes encode different hemagglutinin and neuraminidase proteins. Based on their serotype, there are 18 known types of hemagglutinin and 11 types of neuraminidase.[23][24] Subtypes of IAV are classified by their combination of H and N proteins. For example, "H5N1" designates an influenza A subtype that has a type-5 hemagglutinin (H) protein and a type-1 neuraminidase (N) protein.[23] Additionally, further variation exists within viral subtypes which may lead to significant differences in behavior.[25]
By definition, the subtyping scheme only takes into account the two outer proteins, not the at least eight proteins internal to the virus.[26] Almost all possible combinations of H (1 through 16) and N (1 through 11) have been isolated from wild birds.[9] H17 and H18 have only been discovered in bats.[27]
Influenza virus nomenclature
[edit]Due to the high variability of the virus, subtyping is not sufficient to uniquely identify a strain of influenza A virus. To unambiguously describe a specific isolate of virus, researchers use the Influenza virus nomenclature,[28] which describes, among other things, the subtype, year, and place of collection. Some examples include:[29]
- A/Rio de Janeiro/62434/2021 (H3N2).[29]
- The starting A indicates that the virus is an influenza A virus.
- Rio de Janeiro indicates the place of collection. 62434 is a laboratory sequence number. 2021 (or just 21) indicates that the sample was collected in 2021. No species is mentioned so by default, the sample was collected from a human.
- (H3N2) indicates the subtype of the virus.
- A/swine/South Dakota/152B/2009 (H1N2).[29]
- This example shows an additional field before the place: swine. It indicates that the sample was collected from a pig.
- A/California/04/2009 A(H1N1)pdm09.[29]
- This example carries an unusual designation in the last part: instead of a usual (H1N1), it uses A(H1N1)pdm09. This was in order to distinguish the Pandemic H1N1/09 virus lineage from older H1N1 viruses.[29]
Structure and genetics
[edit]
Structure
[edit]The influenza A virus has a negative-sense, single-stranded, segmented RNA genome, enclosed in a lipid envelope. The virus particle (also called the "virion") is 80–120 nanometers in diameter, such that the smallest virions adopt an elliptical shape; larger virions have a filamentous shape.[30]
Core – The central core of the virion contains the viral RNA genome, which is made of eight separate segments.[31] The nucleoprotein (NP) coats the viral RNA to form a ribonucleoprotein that assumes a helical (spiral) configuration. Three large proteins (PB1, PB2, and PA), which are responsible for RNA transcription and replication, are bound to each segment of viral RNP.[31][32][33]
Capsid – The matrix protein M1 forms a layer between the nucleoprotein and the envelope, called the capsid.[31][32][33]
Envelope – The viral envelope consists of a lipid bilayer derived from the host cell. Two viral proteins; hemagglutinin (HA) and neuraminidase (NA), are inserted into the envelope and are exposed as spikes on the surface of the virion. Both proteins are antigenic; a host's immune system can react to them and produce antibodies in response. The M2 protein forms an ion channel in the envelope and is responsible for uncoating the virion once it has bound to a host cell.[31][32][33]
Genome
[edit]The table below presents a concise summary of the influenza genome and the principal functions of the proteins which are encoded. Segments are conventionally numbered from 1 to 8 in descending order of length.[34][35][36][37]
| RNA segment | Length | Protein | Function |
| 1- PB2 | 2341 | PB2 (Polymerase Basic 2) | A component of the viral RNA polymerase.
PB2 also inhibits JAK1/STAT signaling to inhibit host innate immune response |
| 2- PB1 | 2341 | PB1 (Polymerase Basic 1) | A component of the viral RNA polymerase.
It also degrades the host cell's mitochondrial antiviral signaling protein |
| PB1-F2 (Polymerase Basic 1-Frame 2) | An accessory protein of most IAVs. Not needed for virus replication and growth, it interferes with the host immune response. | ||
| 3- PA | 2233 | PA (Polymerase Acid) | A component of the viral RNA polymerase |
| PA-X | Arises from a ribosomal frameshift in the PA segment. Inhibits innate host immune responses, such as cytokine and interferon production. | ||
| 4- HA | 1775 | HA (Hemagglutinin) | Part of the viral envelope, a protein that binds the virion to host cells, enabling the virus's RNA genetic material to invade it |
| 5- NP | 1565 | NP (Nucleoprotein) | The nucleoprotein associates with the viral RNA to form a ribonucleoprotein (RNP).
At the early stage of infection, the RNP binds to the host cell's importin-α which transports it into the host cell nucleus, where the viral RNA is transcribed and replicated. At a later stage of infection, newly manufactured viral RNA segments assemble with the NP protein and polymerase (PB1, PB2 and PA) to form the core of a progeny virion |
| 6- NA | 1409 | NA (Neuraminidase) | Part of the viral envelope. NA enables the newly assembled virions to escape the host cell and go on to propagate the infection.
NA also facilitates the movement of infective virus particles through mucus, enabling them to reach host epithelial cells. |
| 7- M | 1027 | M1 (Matrix Protein 1) | Forms the capsid, which coats the viral nucleoproteins and supports the structure of the viral envelope.
M1 also assists with the function of the NEP protein. |
| M2 (Matrix Protein 2) | Forms a proton channel in the viral envelope, which is activated once a virion has bound to a host cell. This uncoats the virus, exposing its infective contents to the cytoplasm of the host cell | ||
| 8- NS | 890 | NS1 (non-structural protein 1) | Counteracts the host's natural immune response and inhibits interferon production. |
| NEP (Nuclear Export Protein, formerly NS2 non-structural protein 2) | Cooperates with the M1 protein to mediate the export of viral RNA copies from nucleus into cytoplasm in the late stage of viral replication |
Three viral proteins - PB1, PB2, and PA – associate to form the RNA-dependent RNA polymerase (RdRp) which functions to transcribe and replicate the viral RNA.
Viral messenger RNA transcription – The RdRp complex transcribes viral mRNAs by using a mechanism called cap-snatching. It consists in the hijacking and cleavage of host capped pre-mRNAs. Host cell mRNA is cleaved near the cap to yield a primer for the transcription of positive-sense viral mRNA using the negative-sense viral RNA as a template.[38] The host cell then transports the viral mRNA into the cytoplasm where ribosomes manufacture the viral proteins.[34][35][36][37]
Replication of the viral RNA – The replication of the influenza virus, unlike most other RNA viruses,[39] takes place in the nucleus and involves two steps. The RdRp first of all transcribes the negative-sense viral genome into a positive-sense complimentary RNA (cRNA), then the cRNAs are used as templates to transcribe new negative-sense vRNA copies. These are exported from the nucleus and assemble near the cell membrane to form the core of new virions.[34][35][36][37]
Epidemiology
[edit]Evolution and history
[edit]
The predominant natural reservoir of influenza viruses is thought to be wild waterfowl.[40] The subtypes of influenza A virus are estimated to have diverged 2,000 years ago. Influenza viruses A and B are estimated to have diverged from a single ancestor around 4,000 years ago, while the ancestor of influenza viruses A and B and the ancestor of influenza virus C are estimated to have diverged from a common ancestor around 8,000 years ago.[41]
Outbreaks of influenza-like disease can be found throughout recorded history. The first probable record is by Hippocrates in 412 BCE.[42] The historian Fujikawa listed 46 epidemics of flu-like illness in Japan between 862 and 1868.[43] In Europe and the Americas, a number of epidemics were recorded through the Middle Ages and up to the end of the 19th century.[42]

In 1918-1919 came the first flu pandemic of the 20th century, known generally as the "Spanish flu", which caused an estimated 20 to 50 million deaths worldwide. It is now known that this was caused by an immunologically novel H1N1 subtype of influenza A.[44] The next pandemic took place in 1957, the "Asian flu", which was caused by a H2N2 subtype of the virus in which the genome segments coding for HA and NA appeared to have derived from avian influenza strains by reassortment, while the remainder of the genome was descended from the 1918 virus.[45] The 1968 pandemic ("Hong Kong flu") was caused by a H3N2 subtype in which the NA segment was derived from the 1957 virus, while the HA segment had been reassorted from an avian strain of influenza.[45]
In the 21st century, a strain of H1N1 flu (since titled "H1N1pdm09") was antigenically very different from previous H1N1 strains, leading to a pandemic in 2009. Because of its close resemblance to some strains circulating in pigs, this became known as "swine flu".[46]
Influenza A virus continues to circulate and evolve in birds and pigs. Almost all possible combinations of H (1 through 16) and N (1 through 11) have been isolated from wild birds.[9] As of June 2024, two particularly virulent IAV strains - H5N1 and H7N9 – are predominant in wild bird populations. These frequently cause outbreaks in domestic poultry, with occasional spillover infections in humans who are in close contact with poultry.[47][48]
Pandemic potential
[edit]Influenza viruses have a relatively high mutation rate that is characteristic of RNA viruses.[49] The segmentation of the influenza A virus genome facilitates genetic recombination by segment reassortment in hosts who become infected with two different strains of influenza viruses at the same time.[50][51] With reassortment between strains, an avian strain which does not affect humans may acquire characteristics from a different strain which enable it to infect and pass between humans – a zoonotic event.[52] It is thought that all influenza A viruses causing outbreaks or pandemics among humans since the 1900s originated from strains circulating in wild aquatic birds through reassortment with other influenza strains.[53][54] It is possible (though not certain) that pigs may act as an intermediate host for reassortment.[55]
Surveillance
[edit]The Global Influenza Surveillance and Response System (GISRS) is a global network of laboratories that monitor the spread of influenza with the aim to provide the World Health Organization with influenza control information and to inform vaccine development.[19] Several millions of specimens are tested by the GISRS network annually through a network of laboratories in 127 countries.[20] As well as human viruses, GISRS also monitors avian, swine, and other potentially zoonotic influenza viruses.
Seasonal flu
[edit]
Flu season is an annually recurring time period characterized by the prevalence of an outbreak of influenza, caused either by Influenza A or by Influenza B. The season occurs during the cold half of the year in temperate regions; November through February in the northern hemisphere and May to October in the southern hemisphere. Flu seasons also exist in the tropics and subtropics, with variability from region to region.[57] Annually, about 3 to 5 million cases of severe illness and 290,000 to 650,000 deaths from seasonal flu occur worldwide.[3]
There are several possible reasons for the winter peak in temperate regions:
- During the winter, people spend more time indoors with the windows sealed, so they are more likely to breathe the same air as someone who has the flu and thus contract the virus.[58]
- Days are shorter during the winter, and lack of sunlight leads to low levels of vitamin D and melatonin, both of which require sunlight for their generation. This compromises our immune systems, which in turn decreases ability to fight the virus.[58]
- The influenza virus may survive better in colder, drier climates, and therefore be able to infect more people.[58]
- Cold air reduces the ability of the nasal membranes to resist infection.[59]
Zoonotic infections
[edit]A zoonosis a disease in a human caused by a pathogen (such as a bacterium, or virus) that has jumped from a non-human to a human.[60][61] Avian and pig influenza viruses can, on rare occasions, transmit to humans and cause zoonotic influenza virus infections; these infections are usually confined to people who have been in close contact with infected animals or material such as infected feces and meat, they do not spread to other humans. Symptoms of these infections in humans vary greatly; some are in asymptomatic or mild while others can cause severe disease, leading to severe pneumonia and death.[62] A wide range of Influenza A virus subtypes have been found to cause zoonotic disease.[62][63]
Zoonotic infections can be prevented by good hygiene, by preventing farmed animals from coming into contact with wild animals, and by using appropriate personal protective equipment.[61]
As of June 2024, there is concern about two subtypes of avian influenza which are circulating in wild bird populations worldwide, H5N1 and H7N9. Both of these have potential to devastate poultry stocks, and both have jumped to humans with relatively high case fatality rates.[63] H5N1 in particular has infected a wide range of mammals and may be adapting to mammalian hosts.[64]
Prevention and treatment
[edit]Vaccine
[edit]As of June 2024, the influenza viruses which circulate widely in humans are IAV subtypes H1N1 and H3N2, together with Influenza B.[65] Annual vaccination is the primary and most effective way to prevent influenza and influenza-associated complications, especially for high-risk groups.[66] Vaccines against the flu are trivalent or quadrivalent, providing protection against the dominant strains of IAV(H1N1) and IAV(H3N2), and one or two influenza B virus strains; the formulation is continually reviewed in order to match the predominant strains in circulation.[67][68]
It is possible to vaccinate poultry and pigs against specific strains of influenza. Vaccination should be combined with other control measures such as infection monitoring, early detection and biosecurity.[69][70][71]
Treatment
[edit]The main treatment for mild influenza is supportive; rest, fluids, and over-the-counter medicines to alleviate symptoms while the body's own immune system works to recover from infection. Antiviral drugs are recommended for those with severe symptoms, or for those who are at risk of developing complications such as pneumonia.[72][3]
Signs and symptoms
[edit]Humans
[edit]
The symptoms of seasonal flu are similar to those of a cold, although usually more severe and less likely to include a runny nose.[76] The onset of symptoms is sudden, and initial symptoms are predominately non-specific: a sudden fever; muscle aches; cough; fatigue; sore throat; headache; difficulty sleeping; loss of appetite; diarrhoea or abdominal pain; nausea and vomiting.[77]
Humans can rarely become infected with strains of avian or swine influenza, usually as a result of close contact with infected animals or contaminated material; symptoms generally resemble seasonal flu but occasionally can be severe, including death.[13][14]
Other animals
[edit]Birds
[edit]Some species of wild aquatic birds act as natural asymptomatic carriers of a large variety of influenza A viruses, which they can spread over large distances in their annual migration.[78] Symptoms of avian influenza vary according to both the strain of virus underlying the infection, and on the species of bird affected. Symptoms of influenza in birds may include swollen head, watery eyes, unresponsiveness, lack of coordination, respiratory distress such as sneezing or gurgling.[79]
Highly pathogenic avian influenza
[edit]Because of the impact of avian influenza on economically important chicken farms, avian virus strains are classified as either highly pathogenic (and therefore potentially requiring vigorous control measures) or low pathogenic. The test for this is based solely on the effect on chickens - a virus strain is highly pathogenic avian influenza if 75% or more of chickens die after being deliberately infected with it, or if it is genetically similar to such a strain. The alternative classification is low pathogenic avian influenza.[80] Classification of a virus strain as either a low or high pathogenic strain is based on the severity of symptoms in domestic chickens and does not predict severity of symptoms in other species. Chickens infected with low pathogenic avian influenza display mild symptoms or are asymptomatic, whereas highly pathogenic avian influenza causes serious breathing difficulties, significant drop in egg production, and sudden death.[81]
Since 2006, the World Organization for Animal Health requires all detections of low pathogenic avian influenza H5 and H7 subtypes to be reported because of their potential to mutate into highly pathogenic strains.[82]
Pigs
[edit]Signs of swine flu in pigs can include fever, depression, coughing (barking), discharge from the nose or eyes, sneezing, breathing difficulties, eye redness or inflammation, and going off feed. Some pigs infected with influenza, however, may show no signs of illness at all. Swine flu subtypes are principally H1N1, H1N2, and H3N2;[83] it is spread either through close contact between animals or by the movement of contaminated equipment between farms.[84] Humans who are in close contact with pigs can sometimes become infected.[85]
Horses
[edit]Equine influenza can affect horses, donkeys, and mules;[86] it has a very high rate of transmission among horses, and a relatively short incubation time of one to three days.[87] Clinical signs of equine influenza include fever, nasal discharge, have a dry, hacking cough, depression, loss of appetite and weakness.[87] EI is caused by two subtypes of influenza A viruses: H7N7 and H3N8, which have evolved from avian influenza A viruses.[88]
Dogs
[edit]Most animals infected with canine influenza A will show symptoms such as coughing, runny nose, fever, lethargy, eye discharge, and a reduced appetite lasting anywhere from 2–3 weeks.[89] There are two different influenza A dog flu viruses: one is an H3N8 virus and the other is an H3N2 virus.[89] The H3N8 strain has evolved from an equine influenza avian virus which has adapted to sustained transmission among dogs. The H3N2 strain is derived from an avian influenza which jumped to dogs in 2004 in either Korea or China.[89] It is likely that the virus persists in both animal shelters and kennels, as well as in farms where dogs are raised for meat production.[90]
Bats
[edit]The first bat flu virus, IAV(H17N10), was first discovered in 2009 in little yellow-shouldered bats (Sturnira lilium) in Guatemala.[91] In 2012 a second bat influenza A virus IAV(H18N11) was discovered in flat-faced fruit-eating bats (Artibeus planirostris) from Peru.[92][93][94] Bat influenza viruses have been found to be poorly adapted to non-bat species.[95]
Research
[edit]Influenza research includes efforts to understand how influenza viruses enter hosts, the relationship between influenza viruses and bacteria, how influenza symptoms progress, and why some influenza viruses are deadlier than others.[96] Past pandemics, and especially the 1918 pandemic, are the subject of much research to understand and prevent flu pandemics.[97][98]
The World Health Organization has published a Research Agenda with five streams:[99]
- Stream 1. Reducing the risk of emergence of pandemic influenza. This stream is entirely focused on preventing and limiting pandemic influenza; this includes research into what characteristics make a strain either mild or deadly, worldwide surveillance of influenza A viruses with pandemic potential, and the prevention and management of potentially zoonotic influenza in domestic and farmed animals.[99]
- Stream 2. Limiting the spread of pandemic, zoonotic and seasonal epidemic influenza. This is more broadly targeted at both pandemic and seasonal influenza, looking at the transmission of the virus between people and the ways in which it can spread globally, as well as the environmental and social factors which affect transmission.[99]
- Stream 3. Minimizing the impact of pandemic, zoonotic, and seasonal epidemic influenza. This is principally concerned with vaccination – improving the effectiveness of vaccines, vaccine technology, as well as the speed with which an effective vaccine can be developed and ways in which vaccines can be manufactured and delivered worldwide.[99]
- Stream 4. Optimizing the treatment of patients. This stream aims to reduce the impact of influenza by looking at methods of treatment, vulnerable groups, genetic predispositions, the interaction of influenza infection with other diseases, and influenza sequelae.[99]
- Stream 5. Promoting the development and application of modern public health tools.[99] Aiming to improve the ways in which public policy can combat influenza; this includes the introduction of new technologies, epidemic and pandemic modelling, and the communication of accurate and trustworthy information to the public.[99]
See also
[edit]References
[edit]- ^ "Current ICTV Taxonomy Release". ICTV. Retrieved 28 February 2025.
- ^ "Taxonomy". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 20 March 2020. Retrieved 19 July 2018.
- ^ a b c d "Influenza (Seasonal)". World Health Organization. Retrieved 22 June 2024.
- ^ Runstadler JA, Puryear W (2020). "A Brief Introduction to Influenza A Virus in Marine Mammals". Animal Influenza Virus. Methods in Molecular Biology (Clifton, N.J.). Vol. 2123. pp. 429–450. doi:10.1007/978-1-0716-0346-8_33. ISBN 978-1-0716-0345-1. ISSN 1940-6029. PMID 32170708.
- ^ a b "Influenza A Subtypes and the Species Affected". U.S. Centers for Disease Control and Prevention (CDC). 13 May 2024. Archived from the original on 17 March 2023. Retrieved 17 June 2024.
- ^ Shao W, Li X, Goraya MU, Wang S, Chen JL (August 2017). "Evolution of Influenza A Virus by Mutation and Re-Assortment". International Journal of Molecular Sciences. 18 (8): 1650. doi:10.3390/ijms18081650. PMC 5578040. PMID 28783091.
- ^ Eisfeld AJ, Neumann G, Kawaoka Y (January 2015). "At the centre: influenza A virus ribonucleoproteins". Nature Reviews. Microbiology. 13 (1): 28–41. doi:10.1038/nrmicro3367. PMC 5619696. PMID 25417656.
- ^ "Influenza Type A Viruses". U.S. Centers for Disease Control and Prevention (CDC). 1 February 2024. Retrieved 3 May 2024.
- ^ a b c "FluGlobalNet – Avian Influenza". science.vla.gov.uk. Retrieved 5 June 2024.
- ^ "Types of Influenza Viruses". U.S. Centers for Disease Control and Prevention (CDC). 30 March 2023. Retrieved 17 June 2024.
- ^ "Avian Influenza Type A Viruses". U.S. Centers for Disease Control and Prevention (CDC). 11 June 2024. Retrieved 17 June 2024.
- ^ "Flu". National Health Service. 23 October 2017. Retrieved 17 June 2024.
- ^ a b "Avian influenza: guidance, data and analysis". GOV.UK. 18 November 2021. Retrieved 9 May 2024.
- ^ a b "Swine influenza in humans". European Centre for Disease Prevention and Control (ECDC). 20 September 2017. Retrieved 17 June 2024.
- ^ Joseph U, Su YC, Vijaykrishna D, Smith GJ (January 2017). "The ecology and adaptive evolution of influenza A interspecies transmission". Influenza and Other Respiratory Viruses. 11 (1): 74–84. doi:10.1111/irv.12412. PMC 5155642. PMID 27426214.
- ^ "Avian influenza (bird flu)". European Medicines Agency. 12 June 2024. Retrieved 18 June 2024.
- ^ "Avian influenza (bird flu) vaccination". UK Government – Department for Environment Food & Rural Affairs. 5 June 2023. Retrieved 18 June 2024.
- ^ "What You Should Know about Flu Antiviral Drugs". U.S. Centers for Disease Control and Prevention (CDC). 20 March 2024. Archived from the original on 5 June 2024. Retrieved 18 June 2024.
- ^ a b Lee K, Fang J (2013). Historical Dictionary of the World Health Organization. Rowman & Littlefield. ISBN 9780810878587.
- ^ a b "70 years of GISRS – the Global Influenza Surveillance & Response System". World Health Organization. 19 September 2022. Retrieved 13 June 2024.
- ^ Masurel N (1969). "Serological characteristics of a "new" serotype of influenza A virus: the Hong Kong strain". Bulletin of the World Health Organization. 41 (3): 461–468. PMC 2427714. PMID 5309456.
- ^ Johnson J, Higgins A, Navarro A, Huang Y, Esper FL, Barton N, et al. (February 2012). "Subtyping influenza A virus with monoclonal antibodies and an indirect immunofluorescence assay". Journal of Clinical Microbiology. 50 (2): 396–400. doi:10.1128/JCM.01237-11. PMC 3264186. PMID 22075584.
- ^ a b "Influenza Type A Viruses and Subtypes". U.S. Centers for Disease Control and Prevention (CDC). 2 April 2013. Archived from the original on 1 June 2021. Retrieved 13 June 2013.
- ^ Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, et al. (October 2013). "New world bats harbor diverse influenza A viruses". PLOS Pathogens. 9 (10): e1003657. doi:10.1371/journal.ppat.1003657. PMC 3794996. PMID 24130481.
- ^ "Influenza Virus Genome Sequencing and Genetic Characterization". U.S. Centers for Disease Control and Prevention (CDC). 27 February 2024. Archived from the original on 27 March 2024. Retrieved 19 June 2024.
- ^ Eisfeld AJ, Neumann G, Kawaoka Y (January 2015). "At the centre: influenza A virus ribonucleoproteins". Nature Reviews. Microbiology. 13 (1): 28–41. doi:10.1038/nrmicro3367. PMC 5619696. PMID 25417656.
- ^ "Influenza A Subtypes and the Species Affected". U.S. Centers for Disease Control and Prevention (CDC). 17 June 2024. Retrieved 18 June 2024.
- ^ "A revision of the system of nomenclature for influenza viruses: a WHO memorandum". Bulletin of the World Health Organization. 58 (4): 585–591. 1980. PMC 2395936. PMID 6969132.
This Memorandum was drafted by the signatories listed on page 590 on the occasion of a meeting held in Geneva in February 1980.
- ^ a b c d e "Technical note: Influenza virus nomenclature". Pan American Health Organization. 11 January 2023. Archived from the original on 10 August 2023. Retrieved 27 May 2024.
- ^ Dadonaite B, Vijayakrishnan S, Fodor E, Bhella D, Hutchinson EC (August 2016). "Filamentous influenza viruses". The Journal of General Virology. 97 (8): 1755–1764. doi:10.1099/jgv.0.000535. PMC 5935222. PMID 27365089.
- ^ a b c d Bouvier NM, Palese P (September 2008). "The biology of influenza viruses". Vaccine. 26 (Suppl 4): D49 – D53. doi:10.1016/j.vaccine.2008.07.039. PMC 3074182. PMID 19230160.
- ^ a b c Shaffer C (7 March 2018). "Influenza A Structure". News-Medical. Retrieved 18 June 2024.
- ^ a b c "Virology of human influenza". World Health Organization. 13 May 2010. Retrieved 19 June 2024.
- ^ a b c Krammer F, Smith GJ, Fouchier RA, Peiris M, Kedzierska K, Doherty PC, et al. (June 2018). "Influenza". Nature Reviews. Disease Primers. 4 (1): 3. doi:10.1038/s41572-018-0002-y. PMC 7097467. PMID 29955068.
- ^ a b c Jakob C, Paul-Stansilaus R, Schwemmle M, Marquet R, Bolte H (September 2022). "The influenza A virus genome packaging network – complex, flexible and yet unsolved". Nucleic Acids Research. 50 (16): 9023–9038. doi:10.1093/nar/gkac688. PMC 9458418. PMID 35993811.
- ^ a b c Dou D, Revol R, Östbye H, Wang H, Daniels R (20 July 2018). "Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement". Frontiers in Immunology. 9: 1581. doi:10.3389/fimmu.2018.01581. PMC 6062596. PMID 30079062.
- ^ a b c Rashid F, Xie Z, Li M, Xie Z, Luo S, Xie L (13 December 2023). "Roles and functions of IAV proteins in host immune evasion". Frontiers in Immunology. 14: 1323560. doi:10.3389/fimmu.2023.1323560. PMC 10751371. PMID 38152399.
- ^ Decroly E, Canard B (June 2017). "Biochemical principles and inhibitors to interfere with viral capping pathways". Current Opinion in Virology. 24: 87–96. doi:10.1016/j.coviro.2017.04.003. PMC 7185569. PMID 28527860.
- ^ Rampersad S, Tennant P (2018). "Replication and Expression Strategies of Viruses". Viruses: 55–82. doi:10.1016/B978-0-12-811257-1.00003-6. ISBN 978-0-12-811257-1. PMC 7158166.
- ^ Mahmoud SM, Alison M, Knobler SL, eds. (2005). "1, The Story of Influenza.". The Threat of Pandemic Influenza: Are We Ready? Workshop Summary. Institute of Medicine (US) Forum on Microbial Threats. Washington (DC): National Academies Press (US).
- ^ Suzuki Y, Nei M (April 2002). "Origin and evolution of influenza virus hemagglutinin genes". Molecular Biology and Evolution. 19 (4). Ocford Academic: 501–509. doi:10.1093/oxfordjournals.molbev.a004105. PMID 11919291.
- ^ a b "The History of Influenza". www.flu.com. Retrieved 20 June 2024.
- ^ Shimizu K (October 1997). "[History of influenza epidemics and discovery of influenza virus]". Nihon Rinsho. Japanese Journal of Clinical Medicine. 55 (10): 2505–2511. PMID 9360364.
- ^ "CDC Archives : 1918 Pandemic (H1N1 virus)". U.S. Centers for Disease Control and Prevention (CDC). 20 March 2019. Retrieved 20 June 2024.
- ^ a b Knobler SL, Mack A, Mahmoud A, Lemon SM, et al. (Institute of Medicine (US) Forum on Microbial Threats) (2005). "The Story of Influenza". The Threat of Pandemic Influenza: Are We Ready? Workshop Summary. National Academies Press (US). Retrieved 20 June 2024.
- ^ "2009 H1N1 Pandemic (H1N1pdm09 virus)". CU.S. Centers for Disease Control and Prevention (CDC). 11 June 2019. Retrieved 21 June 2024.
- ^ "The next pandemic: H5N1 and H7N9 influenza?". Gavi, the Vaccine Alliance. 26 March 2021. Retrieved 21 June 2024.
- ^ "Influenza (Avian and other zoonotic)". World Health Organization. 3 October 2023. Retrieved 21 June 2024.
- ^ Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R (October 2010). "Viral mutation rates". Journal of Virology. 84 (19): 9733–9748. doi:10.1128/JVI.00694-10. PMC 2937809. PMID 20660197.
- ^ Kou Z, Lei FM, Yu J, Fan ZJ, Yin ZH, Jia CX, et al. (December 2005). "New genotype of avian influenza H5N1 viruses isolated from tree sparrows in China". Journal of Virology. 79 (24): 15460–15466. doi:10.1128/JVI.79.24.15460-15466.2005. PMC 1316012. PMID 16306617.
- ^ The World Health Organization Global Influenza Program Surveillance Network (October 2005). "Evolution of H5N1 avian influenza viruses in Asia". Emerging Infectious Diseases. 11 (10): 1515–1521. doi:10.3201/eid1110.050644. PMC 3366754. PMID 16318689. Figure 1 shows a diagramatic representation of the genetic relatedness of Asian H5N1 hemagglutinin genes from various isolates of the virus
- ^ "Transmission of Bird Flu Viruses Between Animals and People". U.S. Centers for Disease Control and Prevention (CDC). 15 May 2024. Retrieved 10 June 2024.
- ^ Taubenberger JK, Morens DM (April 2010). "Influenza: the once and future pandemic". Public Health Reports. 125 (Suppl 3): 16–26. doi:10.1177/00333549101250S305. PMC 2862331. PMID 20568566.
- ^ Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (March 1992). "Evolution and ecology of influenza A viruses". Microbiological Reviews. 56 (1): 152–179. doi:10.1128/mr.56.1.152-179.1992. PMC 372859. PMID 1579108.
- ^ "Factsheet on swine influenza in humans and pigs". European Centre for Disease Control. 15 June 2017. Retrieved 13 June 2024.
- ^ CDC U.S. influenza season summary with weekly updates See section 'Pneumonia and Influenza (P&I) Mortality Surveillance' www.cdc.gov, accessed 30 September 2020
- ^ Hirve S, Newman LP, Paget J, Azziz-Baumgartner E, Fitzner J, Bhat N, et al. (27 April 2016). "Influenza Seasonality in the Tropics and Subtropics – When to Vaccinate?". PLOS ONE. 11 (4): e0153003. Bibcode:2016PLoSO..1153003H. doi:10.1371/journal.pone.0153003. PMC 4847850. PMID 27119988.
- ^ a b c "The Reason for the Season: why flu strikes in winter". Science in the News, a Graduate Student Group at the Harvard Graduate School of the Arts and Sciences. 1 December 2014. Retrieved 21 June 2024.
- ^ LaMotte S (6 December 2022). "Scientists finally know why people get more colds and flu in winter". CNN. Retrieved 21 June 2024.
- ^ "zoonosis". Merriam-Webster.com Dictionary. Merriam-Webster. Retrieved 29 March 2019.
- ^ a b "Zoonoses - Key Facts". World Health Organization. 29 July 2020. Retrieved 24 June 2024.
- ^ a b "Zoonotic influenza - Annual Epidemiological Report for 2022". www.ecdc.europa.eu. 23 May 2023. Retrieved 24 June 2024.
- ^ a b "Global AIV with Zoonotic Potential". The Food and Agriculture Organization (FAO) of the United Nations. 29 July 2020. Retrieved 24 June 2024.
- ^ Plaza PI, Gamarra-Toledo V, Euguí JR, Lambertucci SA (March 2024). "Recent Changes in Patterns of Mammal Infection with Highly Pathogenic Avian Influenza A(H5N1) Virus Worldwide". Emerging Infectious Diseases. 30 (3): 444–452. doi:10.3201/eid3003.231098. PMC 10902543. PMID 38407173.
- ^ "Types of Influenza Viruses". U.S. Centers for Disease Control and Prevention (CDC). 30 March 2023. Retrieved 22 June 2024.
- ^ Chow EJ, Doyle JD, Uyeki TM (June 2019). "Influenza virus-related critical illness: prevention, diagnosis, treatment". Critical Care. 23 (1): 214. doi:10.1186/s13054-019-2491-9. PMC 6563376. PMID 31189475.
- ^ Dharmapalan D (October 2020). "Influenza". Indian Journal of Pediatrics. 87 (10): 828–832. doi:10.1007/s12098-020-03214-1. PMC 7091034. PMID 32048225.
- ^ Sautto GA, Kirchenbaum GA, Ross TM (January 2018). "Towards a universal influenza vaccine: different approaches for one goal". Virology Journal. 15 (1): 17. doi:10.1186/s12985-017-0918-y. PMC 5785881. PMID 29370862.
- ^ "Vaccination of poultry against highly pathogenic avian influenza – Available vaccines and vaccination strategies". efsa.europa.eu. 10 October 2023. Retrieved 9 May 2024.
- ^ "Making a Candidate Vaccine Virus (CVV) for a HPAI (Bird Flu) Virus". U.S. Centers for Disease Control and Prevention (CDC). 3 June 2024. Retrieved 15 June 2024.
- ^ "What People Who Raise Pigs Need To Know About Influenza (Flu)". U.S. Centers for Disease Control and Prevention (CDC). 19 October 2023. Retrieved 22 June 2024.
- ^ "Take everyday precautions to protect others while sick". U.S. Centers for Disease Control and Prevention (CDC). 22 March 2024. Retrieved 22 June 2024.
- ^ "Flu Symptoms & Diagnosis". U.S. Centers for Disease Control and Prevention (CDC). 10 July 2019. Archived from the original on 27 December 2019. Retrieved 24 January 2020.
- ^ "Flu Symptoms & Complications". U.S. Centers for Disease Control and Prevention. 26 February 2019. Archived from the original on 1 August 2020. Retrieved 6 July 2019.
- ^ Call SA, Vollenweider MA, Hornung CA, Simel DL, McKinney WP (February 2005). "Does this patient have influenza?". JAMA. 293 (8): 987–997. doi:10.1001/jama.293.8.987. PMID 15728170.
- ^ "Cold Versus Flu". U.S. Centers for Disease Control and Prevention (CDC). 29 September 2022. Retrieved 25 June 2024.
- ^ "Flu". National Health Service UK. 9 August 2023. Retrieved 25 June 2024.
- ^ "Bird flu (avian influenza): how to spot and report it in poultry or other captive birds". Department for Environment, Food & Rural Affairs and Animal and Plant Health Agency. 13 December 2022. Retrieved 6 May 2024.
- ^ "Avian flu". The Royal Society for the Protection of Birds (RSPB). Retrieved 25 June 2024.
- ^ Alexander DJ, Brown IH (April 2009). "History of highly pathogenic avian influenza". Revue Scientifique et Technique. 28 (1): 19–38. doi:10.20506/rst.28.1.1856. PMID 19618616.
- ^ "Avian Influenza in Birds". U.S. Centers for Disease Control and Prevention (CDC). 14 June 2022. Retrieved 6 May 2024.
- ^ "National H5/H7 Avian Influenza surveillance plan". United States Department of Agriculture. Animal Plant Health Inspection Service. October 2013.
- ^ "Factsheet on swine influenza in humans and pigs". European Centre for Disease Prevention and Control. 15 June 2017. Retrieved 25 June 2024.
- ^ "Key Facts about Swine Influenza (Swine Flu) in Pigs". U.S. Centers for Disease Control and Prevention (CDC). 3 October 2018. Retrieved 25 June 2024.
- ^ "2023: outbreaks of swine influenza". World Health Organization. 30 March 2024. Retrieved 25 June 2024.
- ^ "Equine influenza". WOAH – World Organisation for Animal Health. Retrieved 25 June 2024.
- ^ a b "Equine Influenza: Respiratory Diseases of Horses: Merck Veterinary Manual". www.merckvetmanual.com. Archived from the original on 15 November 2016. Retrieved 4 December 2016.
- ^ "Horse Flu". U.S. Centers for Disease Control and Prevention (CDC). 5 May 2023. Retrieved 25 June 2024.
- ^ a b c "Key Facts about Canine Influenza (Dog Flu)". U.S. Centers for Disease Control and Prevention (CDC). 29 August 2023. Retrieved 25 June 2024.
- ^ Wasik BR, Voorhees IE, Parrish CR (January 2021). "Canine and Feline Influenza". Cold Spring Harbor Perspectives in Medicine. 11 (1): a038562. doi:10.1101/cshperspect.a038562. PMC 7778219. PMID 31871238.
- ^ "Bat Influenza (Flu)". cdc.gov. Retrieved 23 March 2025.
- ^ "Characterization of bat influenza viruses". uniklinik-freiburg.de. Retrieved 30 June 2020.
- ^ "New flu virus found in bats". Nature. 503 (7475): 169. 2013. doi:10.1038/503169e. Retrieved 30 June 2020.
- ^ Ciminski K, Pfaff F, Beer M, Schwemmle M (April 2020). "Bats reveal the true power of influenza A virus adaptability". PLOS Pathogens. 16 (4): e1008384. doi:10.1371/journal.ppat.1008384. PMC 7161946. PMID 32298389.
- ^ Ciminski K, Ran W, Gorka M, Lee J, Malmlov A, Schinköthe J, et al. (December 2019). "Bat influenza viruses transmit among bats but are poorly adapted to non-bat species". Nature Microbiology. 4 (12): 2298–2309. doi:10.1038/s41564-019-0556-9. PMC 7758811. PMID 31527796. S2CID 202580293.
- ^ "Influenza Basic Research". National Institute of Allergy and Infectious Diseases. 13 March 2017. Archived from the original on 10 June 2024. Retrieved 24 March 2021.
- ^ Potter CW (October 2001). "A history of influenza". Journal of Applied Microbiology. 91 (4): 572–579. doi:10.1046/j.1365-2672.2001.01492.x. PMID 11576290. S2CID 26392163.
- ^ Taubenberger JK, Baltimore D, Doherty PC, Markel H, Morens DM, Webster RG, et al. (November 2012). "Reconstruction of the 1918 influenza virus: unexpected rewards from the past". mBio. 3 (5). doi:10.1128/mBio.00201-12. PMC 3448162. PMID 22967978.
- ^ a b c d e f g "WHO public health research agenda for influenza: 2017 update". World Health Organization. Geneva. 2017. Retrieved 28 June 2024.
Further reading
[edit]- Official sources
- Pandemic Influenza US CDC
- General information
- Web focus: Warnings of a Flu Pandemic Nature
- Nature Reports: Homepage: Avian Flu
- Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, et al. (September 2005). "Avian influenza A (H5N1) infection in humans". The New England Journal of Medicine. 353 (13): 1374–1385. CiteSeerX 10.1.1.730.7890. doi:10.1056/NEJMra052211. PMID 16192482.
- Pandemic Influenza: Domestic Preparedness Efforts Congressional Research Service Report on Pandemic Preparedness.
- Mahmoud (2005). Stacey L. Knobler, Alison Mack, Mahmoud A, Stanley M. Lemon (eds.). The threat of pandemic influenza : are we ready? : workshop summary / prepared for Forum on Microbial Threats, Board on Global Health. The National Academies Press. p. 285. ISBN 0-309-09504-2.
Highly pathogenic avian influenza virus is on every top ten list available for potential agricultural bioweapon agents
- Mahmoud AA, Institute of Medicine, Knobler S, Mack A (2005). The Threat of Pandemic Influenza: Are We Ready?: Workshop Summary. Washington, D.C.: National Academies Press. ISBN 978-0-309-09504-4.
- Links to Bird Flu pictures (Hardin MD/Univ of Iowa)
- Kawaoka Y (2006). Influenza Virology: Current Topics. Caister Academic Pr. ISBN 978-1-904455-06-6.
- Sobrino F, Mettenleiter T (2008). Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6.
 KSF
KSF