Gene Regulation in Eukaryotes
 From Wikiversity - Reading time: 3 min
From Wikiversity - Reading time: 3 min
Readings
[edit | edit source]- Study guide:
- Wikipedia article: Promoter
- Wikipedia article: Methylation
- Wikipedia article: Imprinting
Control of Eukaryotic Gene Expression
[edit | edit source]Control of eukaryotic gene expression is much more complicated than that of prokaryotic gene expression. This provides more opportunities for regulation. Unlike prokaryotic gene expression, eukaryotic gene expression allows only some genes to be expressed as it does not waste energy this way.
Structure of Eukaryotic Gene
[edit | edit source]Major Differences from Prokaryotic Genes
[edit | edit source]- Exons and introns
- Signal for polyA addition
- More complicated and usually much larger cis regulatory regions surrounding the gene
- Promoter is near the start of transcription
- Enhancer elements are upstream, downstream, or within genes
Promoter
[edit | edit source]The promoter is a cis-acting DNA sequence near the beginning of transcription and usually includes TATA homology. It binds to general transcriptional factors: TFIID, a large complex made up of TATA binding protein (TBP) and TBP associated factors (TAFs).
Enhancer
[edit | edit source]The enhancer is a cis-acting sequence that provides specific activation of nearby promoters and can bind several transcriptional activators or repressors. Distance of the activator/repressor from the promoter region is irrelevant. Enhancers act by looping so that enhancer-bound proteins can interact with promoter-associated proteins. Looping allows multiple enhancers to regulate the expression of a single gene and also allows a single enhancer to regulate more than one promoter or more than one gene, allowing the gene to be independently expressed in response to different stimuli or during different times in development.
Transcription Factors
[edit | edit source]Transcription factors are modular: the DNA binding domains are usually separate from activation or repressed domains. They often act as dimers binding to a palindromic or nearly palindromic site.
Signaling Pathways
[edit | edit source]Signaling specificity depends on three principles of transcriptional control: (1) activator insufficiency, (2) default repression, and (3) cooperative activation. These interconnected transcriptional control principles place tight restrictions on the expression of target genes, but allow robust expression under appropriate stimulus.
Activator Insufficiency
[edit | edit source]Activator insufficiency is the inability of signal-regulated transcription factor alone to strongly activate gene expression. It prevents signaling pathways from activating all of their target genes during each signaling event.
Activator insufficiency prevents promiscuous activation.
Default Repression
[edit | edit source]Default repression is mediated by signal-stimulating transcription factors in the absence of signaling and limits target gene expression to cells that both express the appropriate local activators and receive signaling.
Cooperative Activation
[edit | edit source]Signal-regulated activators must act cooperatively with tissue-specific or cell-specific local activators to drive high levels of target gene expression.
Epigenetic Regulation
[edit | edit source]Epigenetic regulations are usually due to chemical modification of DNA bases or protein complexes stably bound to DNA. These changes are heritable and are not due to changes in the DNA sequence itself.
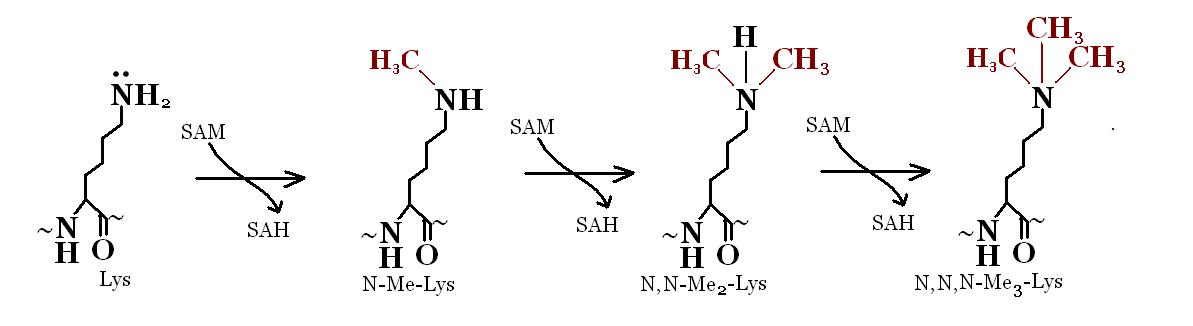
DNA methylation
[edit | edit source]In many higher eukaryotes, cytosine is methylated at carbon 5 by DNA methylase enzyme. In mammals, the methylated sequence is usually C*pG. Inactive genes are preferentially methylated. Active genes are hypomethylated.
This methylation does not change the nucleotide sequence but can be propogated at DNA replication. Methylated DNA can be detected by two restriction enzymes that recognize CCGG: MspI which is methylation insensitive and HpaII which is methylation sensitive.
Imprinting
[edit | edit source]In mammals a few clustered genes are turned off in the germline of one parent. Once imprinted, a gene stays off throughout embryogenesis and adult life in somatic cells. The imprint is removed early in germline development and then re-established in sex-specific patterns.
The individuals with dots in them represent individuals who carried the gene but did not show mutant phenotype.
Prader-Willi and Angelman Syndromes
[edit | edit source]3 imprinted genes control Prader-Willi and Angelman Syndromes: SNRPN. necdin, and Ube3. For females, SNRPN and necdin are off while Ube3 is in. For males, SNRPN and necdin are on while Ube3 is off. Children inheriting a deletion from their father will have no active SNRPN or necdin genes and will show Prader-Willi syndrom. Children inheriting a deletion from their mother will have no active Ube3 gene and will show Angelman syndrome.
See also
[edit | edit source]- RNA interference - 2006 Nobel Prize to Andrew Z. Fire and Craig C. Mello
- Eukaryotic transcription - 2006 Nobel Prize to Roger D. Kornberg
External links
[edit | edit source]- Gene regulation book modules at Wikibooks need help.
- Modern Genetic Analysis by Anthony J.F. Griffiths, William M. Gelbart, Jeffrey H. Miller and Richard C. Lewontin (1999) published by W. H. Freeman & Co.
 KSF
KSF