Protein Crystallography project
 From Wikiversity - Reading time: 8 min
From Wikiversity - Reading time: 8 min
This is a Protein Crystallography Project concerned with the Determination of Protein Structures in Crystals and in Solutions with electrolytes by X-ray Diffraction, neutron diffraction, light scattering, Vibrational Circular Dichroism (VCD) and 2D-NMR.
Course on Protein Biochemistry
[edit | edit source]Course and Lecture Notes on X-ray Diffraction by Protein Crystals
[edit | edit source]
Course and Lecture Notes on Neutron Diffraction by Protein Crystals
[edit | edit source]Course and Lecture Notes on 2D-NMR Determination of Protein Structure in Solution
[edit | edit source]Lecture Notes of NMR on Magnetically-oriented Protein Microcrystals in suspension
[edit | edit source]
Course on Computer Modeling of Protein Structure and Dynamics
[edit | edit source]Lecture Notes on Paracrystal Theory and Quasi-crystals
[edit | edit source]- The following figures and data have the following Source: An Introduction to Molecular Biology/Function and structure of Proteins


Amino Acid Structures and Properties
[edit | edit source]The 20 amino acids encoded directly by the genetic code can be divided into several groups based on their properties. Important factors are charge, hydrophilicity or hydrophobicity, size and functional groups.Amino acids are usually classified by the properties of their side chain into four groups. The side chain can make an amino acid a weak acid or a weak base, and a hydrophile if the side chain is polar or a hydrophobe if it is nonpolar.

Protein amino acids are combined into a single polypeptide chain in a condensation reaction. This reaction is catalysed by the ribosome in a process known as translation.
| Essential | Nonessential |
|---|---|
| Isoleucine | Alanine |
| Leucine | Asparagine |
| Lysine | Aspartic Acid |
| Methionine | Cysteine* |
| Phenylalanine | Glutamic Acid |
| Threonine | Glutamine* |
| Tryptophan | Glycine* |
| Valine | Proline* |
| Selenocysteine* | |
| Serine* | |
| Tyrosine* | |
| Arginine* | |
| Histidine* | |
| Ornithine* | |
| Taurine* |
Polar and non polar amino acids and their single and three letter code
| Amino Acid | Three Letter code | Single Letter code | Side chain polarity | Side chain charge (pH 7.4) | Hydropathy index | Absorbance λmax(nm) | ε at λmax (x10−3 M−1 cm−1) |
|---|---|---|---|---|---|---|---|
| Alanine | Ala | A | nonpolar | neutral | 1.8 | ||
| Arginine | Arg | R | polar | positive | −4.5 | ||
| Asparagine | Asn | N | polar | neutral | −3.5 | ||
| Aspartic acid | Asp | D | polar | negative | −3.5 | ||
| Cysteine | Cys | C | nonpolar | neutral | 2.5 | 250 | 0.3 |
| Glutamic acid | Glu | E | polar | negative | −3.5 | ||
| Glutamine | Gln | Q | polar | neutral | −3.5 | ||
| Glycine | Gly | G | nonpolar | neutral | −0.4 | ||
| Histidine | His | H | polar | positive(10%)
neutral(90%) |
−3.2 | 211 | 5.9 |
| Isoleucine | Ile | I | nonpolar | neutral | 4.5 | ||
| Leucine | Leu | L | nonpolar | neutral | 3.8 | ||
| Lysine | Lys | K | polar | positive | −3.9 | ||
| Methionine | Met | M | nonpolar | neutral | 1.9 | ||
| Phenylalanine | Phe | F | nonpolar | neutral | 2.8 | 257, 206, 188 | 0.2, 9.3, 60.0 |
| Proline | Pro | P | nonpolar | neutral | −1.6 | ||
| Serine | Ser | S | polar | neutral | −0.8 | ||
| Threonine | Thr | T | polar | neutral | −0.7 | ||
| Tryptophan | Trp | W | nonpolar | neutral | −0.9 | 280, 219 | 5.6, 47.0 |
| Tyrosine | Tyr | Y | polar | neutral | −1.3 | 274, 222, 193 | 1.4, 8.0, 48.0 |
| Valine | Val | V | nonpolar | neutral | 4.2 |
Additionally, there are two additional amino acids which are incorporated by overriding stop codons:
| 21st and 22nd amino acids | 3-Letter | 1-Letter |
|---|---|---|
| Selenocysteine | Sec | U |
| Pyrrolysine | Pyl | O |
In addition to the specific amino acid codes, placeholders are used in cases where chemical or crystallographic analysis of a peptide or protein can not conclusively determine the identity of a residue.
| Ambiguous Amino Acids | 3-Letter | 1-Letter |
|---|---|---|
| Asparagine or aspartic acid | Asx | B |
| Glutamine or glutamic acid | Glx | Z |
| Leucine or Isoleucine | Xle | J |
| Unspecified or unknown amino acid | Xaa | X |
Amino Acid Structure Gallery
[edit | edit source]-
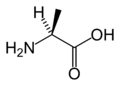
L-Alanine
(Ala / A) -
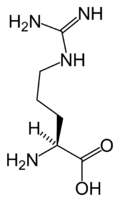
L-Arginine
(Arg / R) -
L-Asparagine
(Asn / N) -
L-Aspartic acid
(Asp / D) -
L-Cysteine
(Cys / C) -
L-Glutamic acid
(Glu / E) -
L-Glutamine
(Gln / Q) -
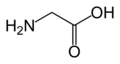
Glycine
(Gly / G) -
L-Histidine
(His / H) -
L-Isoleucine
(Ile / I) -
L-Leucine
(Leu / L) -
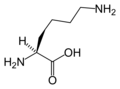
L-Lysine
(Lys / K) -
L-Methionine
(Met / M) -
L-Phenylalanine
(Phe / F) -
L-Proline
(Pro / P) -
L-Serine
(Ser / S) -
L-Threonine
(Thr / T) -
L-Tryptophan
(Trp / W) -
L-Tyrosine
(Tyr / Y) -
L-Valine
(Val / V) -
L-Selenocysteine
(Sec / U) -
L-Pyrrolysine
(Pyl / O)
The 20 naturally occurring amino acids have different physical and chemical properties, including their electrostatic charge, pKa, hydrophobicity, size and specific functional groups. These properties play a major role in molding protein structure. The salient features of amino acids are described below in the table.
| Amino Acid | Abbrev. | Remarks | |
|---|---|---|---|
Alanine |
A | Ala | Very abundant, very versatile. More stiff than glycine, but small enough to pose only small steric limits for the protein conformation. It behaves fairly neutrally, and can be located in both hydrophilic regions on the protein outside and the hydrophobic areas inside. |
| Asparagine or aspartic acid | B | Asx | A placeholder when either amino acid may occupy a position. |
Cysteine |
C | Cys | The sulfur atom bonds readily to heavy metal ions. Under oxidizing conditions, two cysteines can join together in a disulfide bond to form the amino acid cystine. When cystines are part of a protein, insulin for example, the tertiary structure is stabilized, which makes the protein more resistant to denaturation; therefore, disulfide bonds are common in proteins that have to function in harsh environments including digestive enzymes (e.g., pepsin and chymotrypsin) and structural proteins (e.g., keratin). Disulfides are also found in peptides too small to hold a stable shape on their own (eg. insulin). |
Aspartic acid |
D | Asp | Behaves similarly to glutamic acid. Carries a hydrophilic acidic group with strong negative charge. Usually is located on the outer surface of the protein, making it water-soluble. Binds to positively-charged molecules and ions, often used in enzymes to fix the metal ion. When located inside of the protein, aspartate and glutamate are usually paired with arginine and lysine. |
Glutamic acid |
E | Glu | Behaves similar to aspartic acid. Has longer, slightly more flexible side chain. |
Phenylalanine |
F | Phe | Essential for humans. Phenylalanine, tyrosine, and tryptophan contain large rigid aromatic group on the side-chain. These are the biggest amino acids. Like isoleucine, leucine and valine, these are hydrophobic and tend to orient towards the interior of the folded protein molecule. Phenylalanine can be converted into Tyrosine. |
Glycine |
G | Gly | Because of the two hydrogen atoms at the α carbon, glycine is not optically active. It is the smallest amino acid, rotates easily, adds flexibility to the protein chain. It is able to fit into the tightest spaces, e.g., the triple helix of collagen. As too much flexibility is usually not desired, as a structural component it is less common than alanine. |
Histidine |
H | His | In even slightly acidic conditions protonation of the nitrogen occurs, changing the properties of histidine and the polypeptide as a whole. It is used by many proteins as a regulatory mechanism, changing the conformation and behavior of the polypeptide in acidic regions such as the late endosome or lysosome, enforcing conformation change in enzymes. However only a few histidines are needed for this, so it is comparatively scarce. |
Isoleucine |
I | Ile | Essential for humans. Isoleucine, leucine and valine have large aliphatic hydrophobic side chains. Their molecules are rigid, and their mutual hydrophobic interactions are important for the correct folding of proteins, as these chains tend to be located inside of the protein molecule. |
| Leucine or isoleucine | J | Xle | A placeholder when either amino acid may occupy a position |
Lysine |
K | Lys | Essential for humans. Behaves similarly to arginine. Contains a long flexible side-chain with a positively-charged end. The flexibility of the chain makes lysine and arginine suitable for binding to molecules with many negative charges on their surfaces. E.g., DNA-binding proteins have their active regions rich with arginine and lysine. The strong charge makes these two amino acids prone to be located on the outer hydrophilic surfaces of the proteins; when they are found inside, they are usually paired with a corresponding negatively-charged amino acid, e.g., aspartate or glutamate. |
Leucine |
L | Leu | Essential for humans. Behaves similar to isoleucine and valine. See isoleucine. |
Methionine |
M | Met | Essential for humans. Always the first amino acid to be incorporated into a protein; sometimes removed after translation. Like cysteine, contains sulfur, but with a methyl group instead of hydrogen. This methyl group can be activated, and is used in many reactions where a new carbon atom is being added to another molecule. |
Asparagine |
N | Asn | Similar to aspartic acid. Asn contains an amide group where Asp has a carboxyl. |
| Pyrrolysine | O | Pyl | Similar to lysine, with a pyrroline ring attached. |
Proline |
P | Pro | Contains an unusual ring to the N-end amine group, which forces the CO-NH amide sequence into a fixed conformation. Can disrupt protein folding structures like α helix or β sheet, forcing the desired kink in the protein chain. Common in collagen, where it often undergoes a posttranslational modification to hydroxyproline. |
Glutamine |
Q | Gln | Similar to glutamic acid. Gln contains an amide group where Glu has a carboxyl. Used in proteins and as a storage for ammonia. The most abundant Amino Acid in the body. |
Arginine |
R | Arg | Functionally similar to lysine. |
Serine |
S | Ser | Serine and threonine have a short group ended with a hydroxyl group. Its hydrogen is easy to remove, so serine and threonine often act as hydrogen donors in enzymes. Both are very hydrophilic, therefore the outer regions of soluble proteins tend to be rich with them. |
Threonine |
T | Thr | Essential for humans. Behaves similarly to serine. |
| Selenocysteine | U | Sec | Selenated form of cysteine, which replaces sulfur. |
Valine |
V | Val | Essential for humans. Behaves similarly to isoleucine and leucine. See isoleucine. |
Tryptophan |
W | Trp | Essential for humans. Behaves similarly to phenylalanine and tyrosine (see phenylalanine). Precursor of serotonin. Naturally fluorescent. |
| Unknown | X | Xaa | Placeholder when the amino acid is unknown or unimportant. |
Tyrosine |
Y | Tyr | Behaves similarly to phenylalanine (precursor to Tyrosine) and tryptophan (see phenylalanine). Precursor of melanin, epinephrine, and thyroid hormones. Naturally fluorescent, although fluorescence is usually quenched by energy transfer to tryptophans. |
| Glutamic acid or glutamine | Z | Glx | A placeholder when either amino acid may occupy a position. |
References
[edit | edit source]↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Protein&oldid=425576197 ↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Proteinogenic_amino_acid&oldid=420804587 ↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Amino_acid&oldid=425389108 ↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Amino_acid&oldid=425389108 ↑ Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells.Nature Methods:4,261–7,2005 ↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Peptide_bond&oldid=417601014 ↑ Basler B, Schuster O, Bach T (November 2005). "Conformationally constrained β-amino acid derivatives by intramolecular [2 + 2]-photocycloaddition of a tetronic acid amide and subsequent lactone ring opening". J. Org. Chem. 70 (24): 9798–808. doi:10.1021/jo0515226. PMID 16292808. ↑ Murray JK, Farooqi B, Sadowsky JD, et al. (September 2005). "Efficient synthesis of a β-peptide combinatorial library with microwave irradiation". J. Am. Chem. Soc. 127 (38): 13271–80. doi:10.1021/ja052733v. PMID 16173757. ↑ Seebach D, Matthews JL (1997). "β-Peptides: a surprise at every turn". Chem. Commun. (21): 2015–22. doi:10.1039/a704933a. ↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Enzyme&oldid=424282616 ↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Enzyme&oldid=424282616 ↑ Moss, G.P.. "Recommendations of the Nomenclature Committee". International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes by the Reactions they Catalyse. Retrieved 2006-03-14. ↑ http://en.wikipedia.orghttps://en.wikiversity.org/w/index.php?title=Enzyme&oldid=424282616 ↑ Lovell SC et al. (2003). "Structure validation by Cα geometry: φ,ψ and Cβ deviation". Proteins 50 (3): 437–450. doi:10.1002/prot.10286. ... ↑ Crowfoot Hodgkin D (1935). "X-ray Single Crystal Photographs of Insulin". Nature 135: 591. doi:10.1038/135591a0. ↑ Kendrew J. C. et al. (1958-03-08). "A Three-Dimensional Model of the Myoglobin Molecule Obtained by X-Ray Analysis". Nature 181 (4610): 662. doi:10.1038/181662a0. PMID 13517261. ↑ "Table of entries in the PDB, arranged by experimental method". ↑ "PDB Statistics". RCSB Protein Data Bank. Retrieved 2010-02-09. ↑ Scapin G (2006). "Structural biology and drug discovery". Curr. Pharm. Des. 12 (17): 2087. doi:10.2174/138161206777585201. PMID 16796557. ↑ Lundstrom K (2006). "Structural genomics for membrane proteins". Cell. Mol. Life Sci. 63 (22): 2597. doi:10.1007/s00018-006-6252-y. PMID 17013556. ↑ Lundstrom K (2004). "Structural genomics on membrane proteins: mini review". Comb. Chem. High Throughput Screen. 7 (5): 431. PMID 15320710.
 KSF
KSF